Visualized detection method of BAR transgenic crops
A detection method and crop technology, applied in the field of genetic engineering, can solve the problem of high detection cost of fluorescent probes, achieve the effects of cheap reagents, lower detection costs, and convenient synthesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
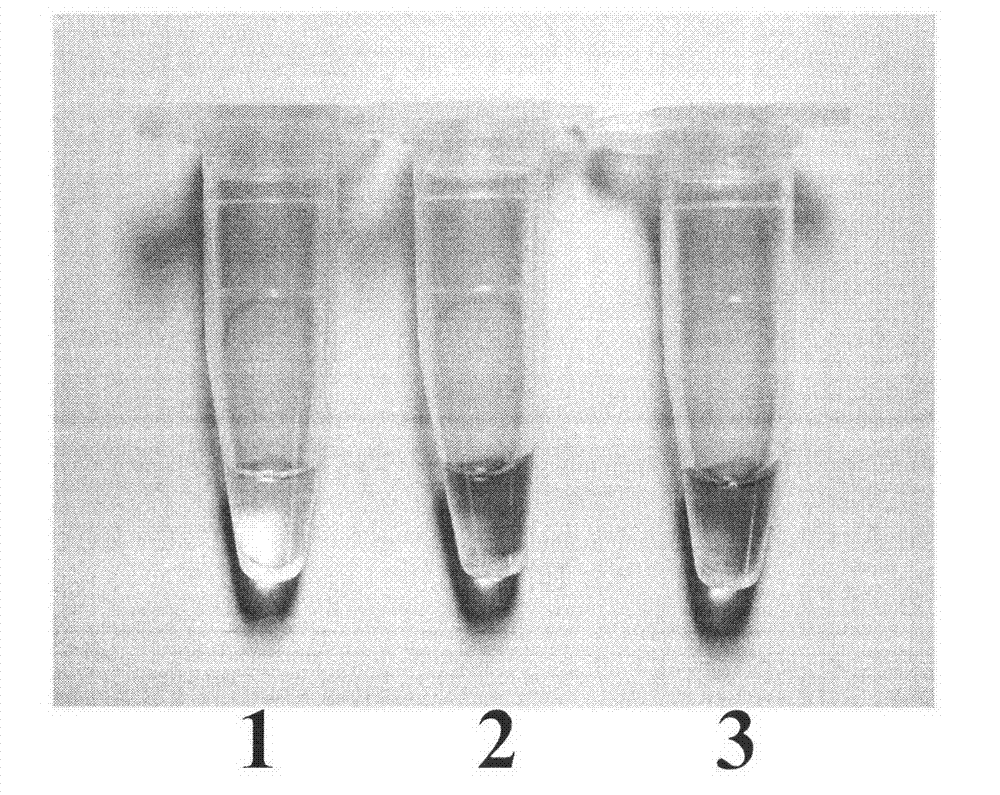
[0020] Embodiment 1: Detection of BAR gene in rice
[0021] 1. Extraction of rice genomic DNA;
[0022] Rice genomic DNA was extracted by the improved CTAB method. ①Grind an appropriate amount of rice leaves or seeds into a fine powder in a mortar under the protection of liquid nitrogen, put a small amount of the powder into a 1.5mL EP tube, add 700μl of CTAB extraction buffer preheated at 65°C, shake and mix well, and incubate at 65°C for 2h ; ② Add 600 μl of chloroform-isoamyl alcohol (24:1) after the sample is cooled to room temperature, and mix it upside down for 5 minutes; ③ Centrifuge at 7500r.min-1 for 10 minutes, and transfer the supernatant to a new EP tube; ④ Add etc. Volume -20 ℃ pre-cooled isopropanol, mix well, and place at 20 ℃ for 30 minutes; ⑤10,000r.min-1 centrifuge for 15 minutes, discard the supernatant; Centrifuge at 10,000r.min-1 for 3 minutes, discard the supernatant, evaporate ethanol at room temperature, dissolve DNA in sterile high-purity water, and ...
Embodiment 2
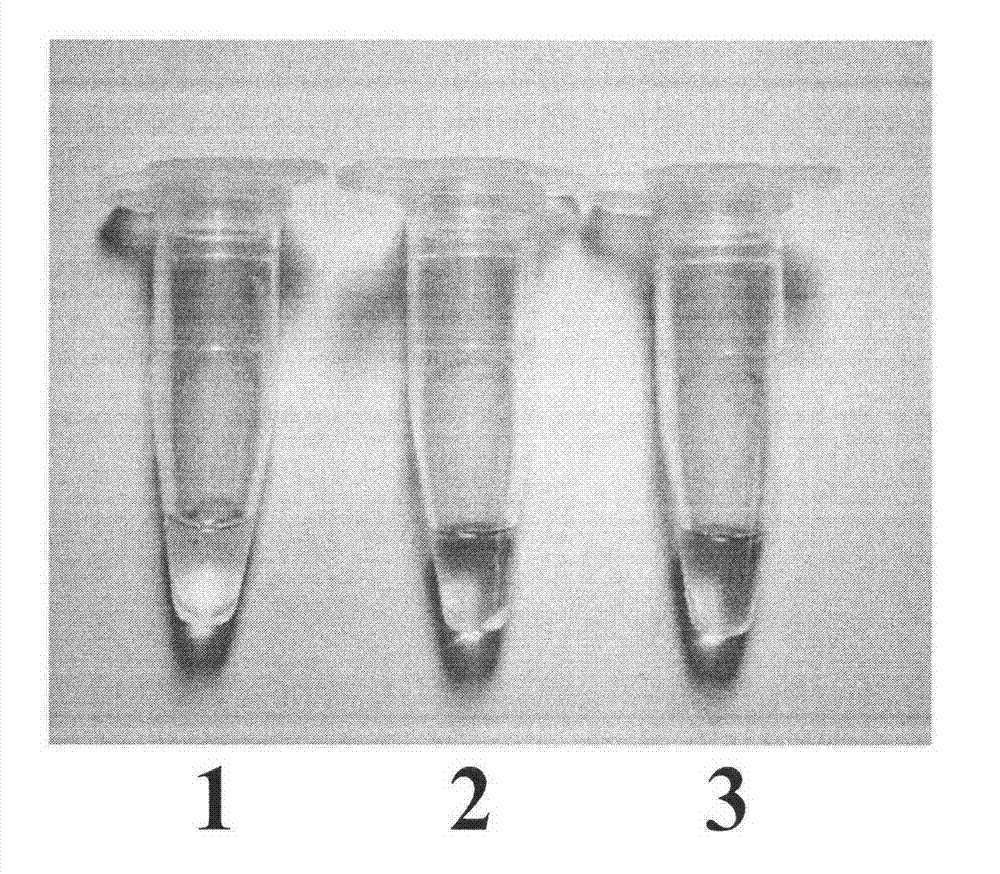
[0033] Embodiment 2: the detection of BAR gene in cotton
[0034] 1. Extraction of cotton genomic DNA;
[0035] Cotton genomic DNA was extracted by the improved CTAB method. ①Grind an appropriate amount of cotton leaves or seeds into a fine powder in a mortar under the protection of liquid nitrogen, take a small amount of powder into a 1.5mL EP tube, add 700μl of CTAB extraction buffer preheated at 65°C, shake and mix well, and incubate at 65°C for 2h ; ② Add 600 μl of chloroform-isoamyl alcohol (24:1) after the sample is cooled to room temperature, and mix it upside down for 5 minutes; ③ Centrifuge at 7500r.min-1 for 10 minutes, and transfer the supernatant to a new EP tube; ④ Add etc. Volume -20 ℃ pre-cooled isopropanol, mix well, and place at 20 ℃ for 30 minutes; ⑤10,000r.min-1 centrifuge for 15 minutes, discard the supernatant; Centrifuge at 10,000r.min-1 for 3 minutes, discard the supernatant, evaporate ethanol at room temperature, dissolve DNA in sterile high-purity wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com