Inhibitor for inducing pluripotent stem cells, and inducing method and application thereof
A multifunctional stem cell and inhibitor technology, applied in the field of pharmaceutical bioengineering, can solve the problems of increasing the risk of iPS clinical application, limiting iPS application, iPS cell carcinogenesis, etc., and achieving good structural stability, ability to pass through cell membrane, and induction efficiency. improved effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
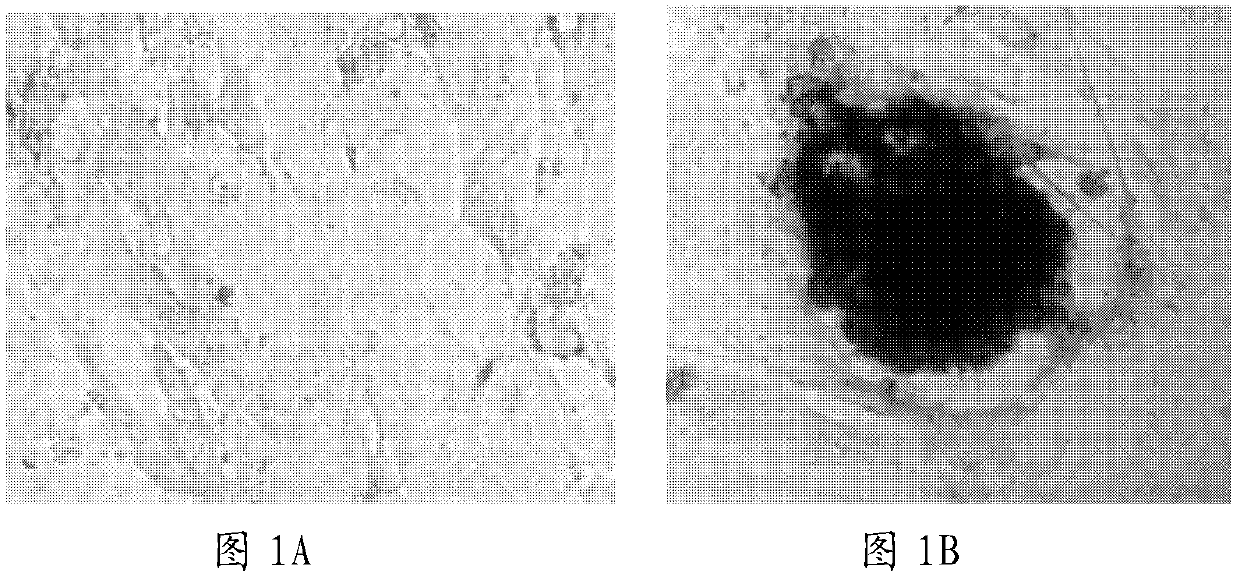
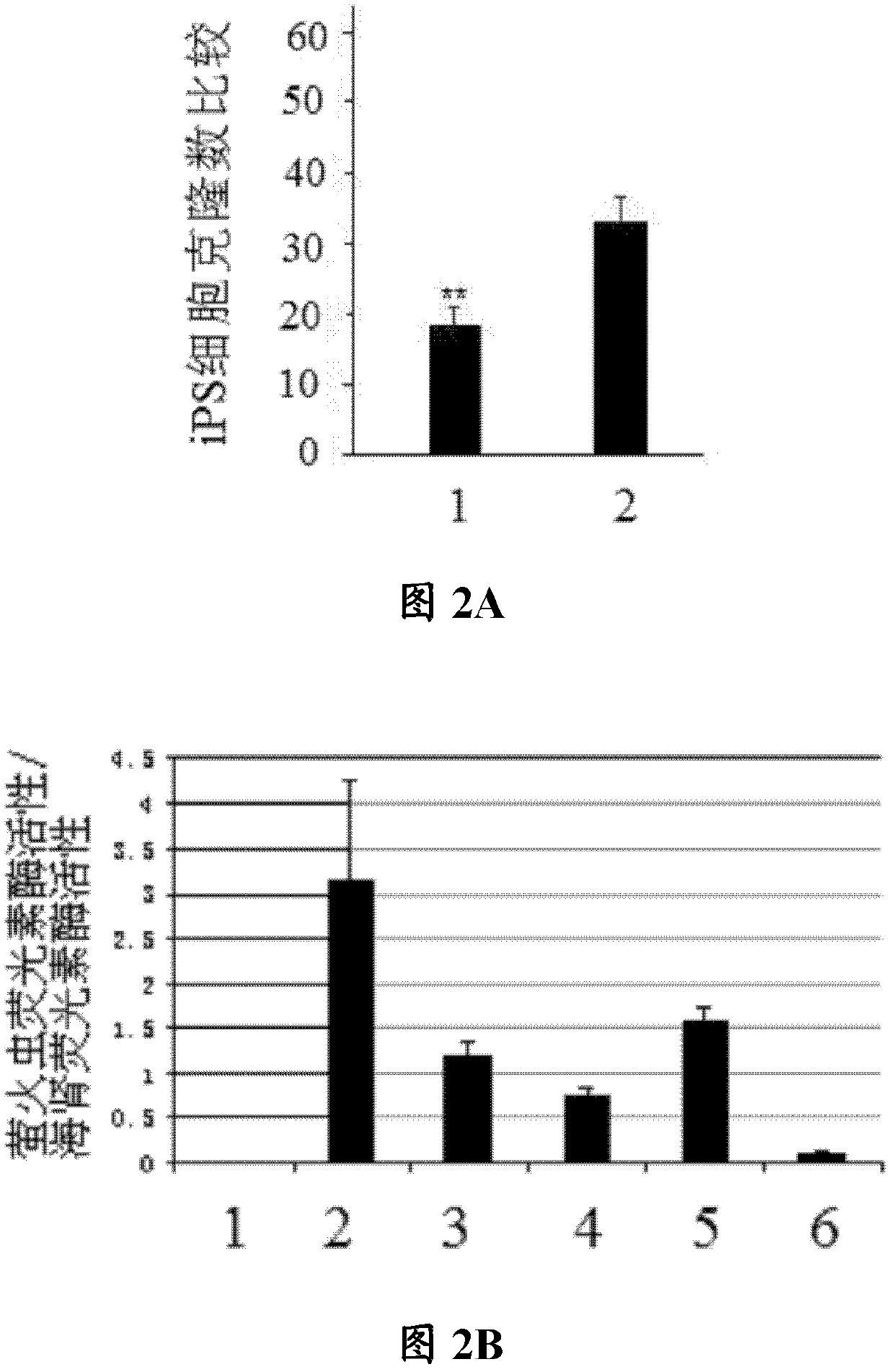
[0059] Example 1 microRNA-421 inhibitor induces iPS cells
[0060] In this example, cardiac fibroblasts were isolated from mouse heart tissue, and microRNA-421 inhibitors were used to induce the cardiac fibroblasts to generate iPS cells. The specific experimental steps are as follows:
[0061] Take an adult mouse, open the chest and cut three hearts under sterile conditions, rinse the heart three times with PBS with pH 7.4, then place it on ice, add 1 mL of DMEM with 0.08% trypsin and 1% collagenase to digest 50mL of DMEM digestion solution containing 2mL of 0.08% trypsin and 2mL of 1% collagenase II (trypsin and collagenase II were ordered from Beijing Xinjingke Biotechnology Co., Ltd. and sigma company respectively). Chop into small pieces of 1-2mm. Then add 5mL of digestion solution (50mL of DMEM digestion solution contains 2mL of 0.08% trypsin and 2mL of 1% collagenase II) for digestion, and at 37°C, digest at a constant temperature in a shaker for 4-6 minutes (the sh...
Embodiment 2
[0066] Example 2 Preparation and identification of mouse model of myocardial infarction
[0067] In this example, a mouse model of myocardial infarction was prepared and identified.
[0068] 1) The method of blocking the left coronary artery is used to induce myocardial infarction in mice, thereby establishing a mouse model of myocardial infarction.
[0069] After anesthesia, the mice were fixed on the experimental wooden board in the dorsal position. ECG monitoring. The trachea was incised in the middle of the neck and intubated, connected to the animal respirator, the chest was opened in the fourth intercostal space, the pericardium was carefully lifted and cut open to fully expose the heart and blood vessels. Between the conus pulmonary artery and the left atrial appendage, with the main trunk of the left coronary vein as the mark, insert the needle below the root of the left atrial appendage with a depth of 0.5mm; pass the 6 / 0 suture through the surface layer of the m...
Embodiment 3
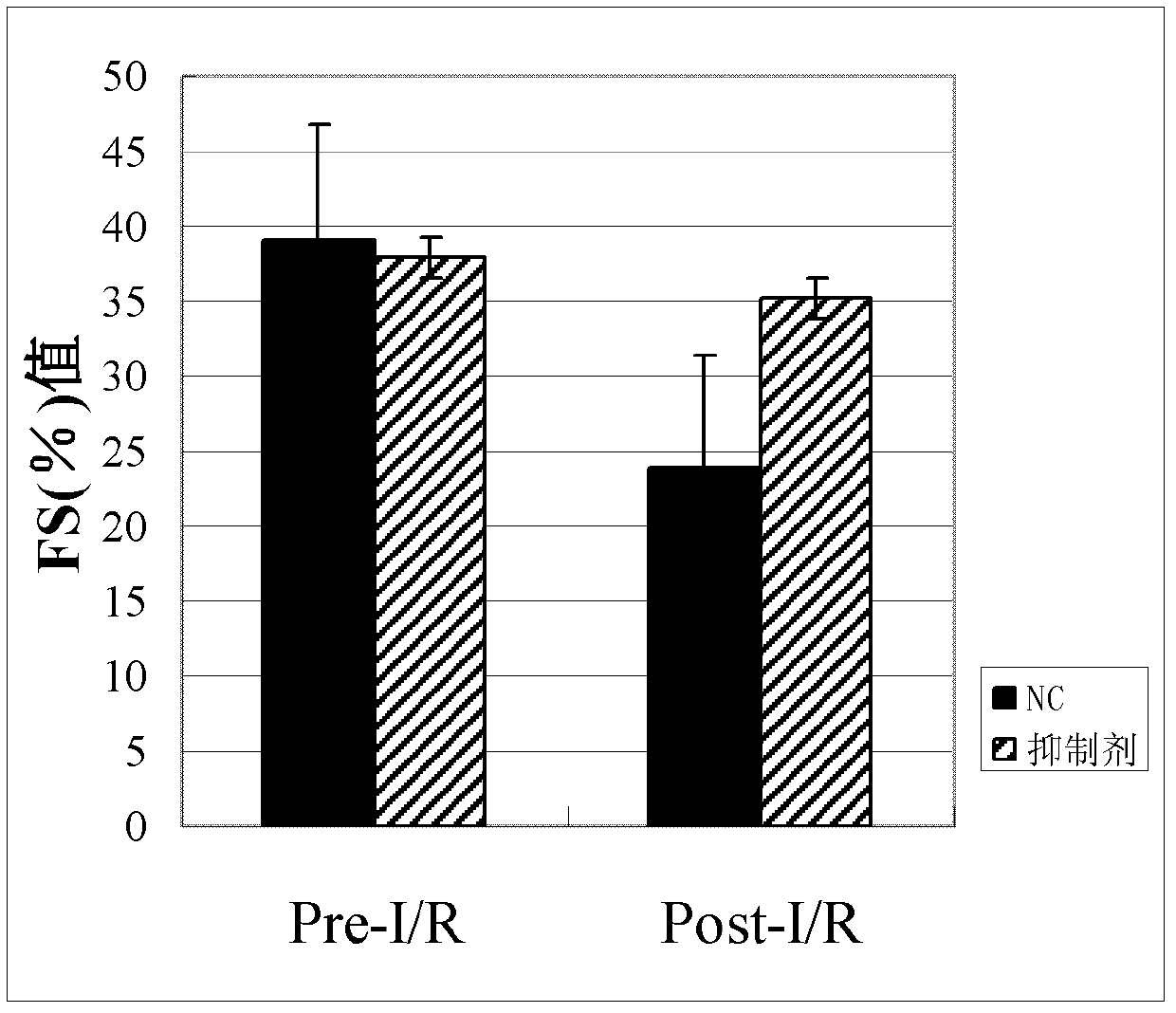
[0084] Example 3 Using mi-iPS cells to treat myocardial infarction in mice, observe the changes in the size of myocardial infarction and the degree of cardiac function repair in mice.
[0085] Six-week-old C57 mice were selected to prepare myocardial infarction model mice according to step (1) of Example 2. With five mice as the control group and five mice as the experimental group, 1 × 10 6 mi-iPS cells. After 1 week and 1 month, the change of the area of myocardial infarction in the mice and the recovery degree of the myocardial function of the mice were detected by using an ultrasonocardiometer. The change (FS% value) of mouse cardiac function after one month sees image 3 .
[0086] image 3 The data presented in are observations after one month.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com