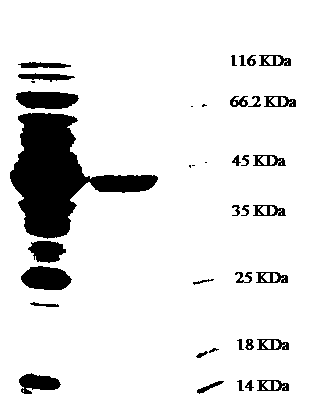
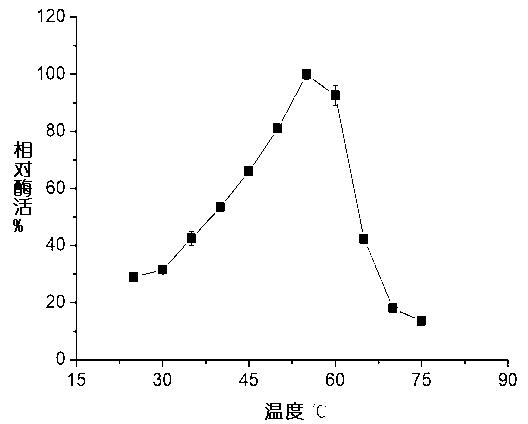
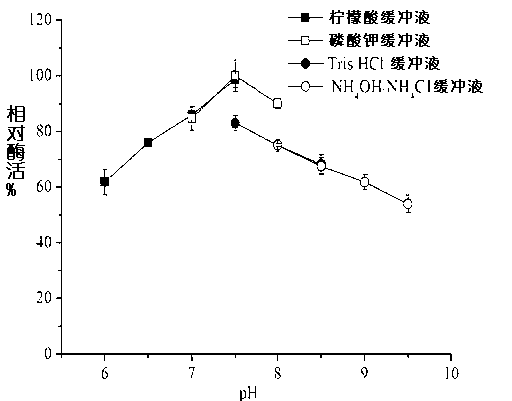
Nitrilase, gene sequence and application method thereof
A nitrilase and sequence technology, applied in the field of enzyme genetic engineering and enzyme engineering, can solve the problems of cloning, expression, and few reports of enzymatic properties, and achieve the advantages of short fermentation period, high catalytic efficiency and high nitrilase activity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] This example illustrates the method for cloning the partial sequence (Nit M) of the nitrilase gene.
[0039] Pseudomonas putida ( Pseudomonas putida ) XY4 (preservation number CGMCC No.3830) cloning of Nit M sequence: the Pseudomonas The amino acid sequence of nitrilase was homologously compared, and the primers were designed by CODEHOP software. After further analysis by DNAMAN software, a pair of degenerate primers P1 and P2 were designed to target Pseudomonas putida ( Pseudomonas putida ) XY4 (Accession No. CGMCC No.3830) The total DNA was used as a template to clone the Nit M sequence:
[0040] P1: 5'-GGCGGAAGCTGaarccnacnca-3'
[0041] P2: 5'-ACGTCGGGCCTGswrtartgncc-3'
[0042] The PCR reaction was carried out in a 50 μL system, and the reaction conditions were pre-denaturation at 95°C for 5 min; denaturation at 94°C for 30 s, annealing at 60°C for 40 s, and extension at 72°C for 40 s, a total of 30 cycles; and final extension at 72°C for 10 min...
Embodiment 2
[0044] This example illustrates the cloning method of the 5' end sequence (Nit 5') of the nitrilase gene.
[0045] Pseudomonas putida ( Pseudomonas putida ) Cloning of the Nit 5' sequence of XY4 (Deposit No. CGMCC No.3830): According to the isolation and purification of Pseudomonas putida ( Pseudomonas putida ) XY4 (preservation number CGMCC No.3830) N-terminal sequence measured by the nitrilase produced by designing degenerate primer P3, and primer P4 was designed according to the Nit M sequence obtained above, and Pseudomonas putida ( Pseudomonas putida ) XY4 (preservation number CGMCC No.3830) total DNA as template for cloning Nit 5' end sequence:
[0046] P3: 5’-ATGGTRACATAYACNAAYAA-3’
[0047] P4: 5'-TCTACATGCGTCGGCTTCAGCTT-3'
[0048] The PCR reaction was carried out in a 50 μL system, and the reaction conditions were pre-denaturation at 95°C for 5 min; denaturation at 94°C for 30 s, annealing at 55°C for 40 s, and extension at 72°C for 40 s, a total of 3...
Embodiment 3
[0050] This example illustrates the cloning method of the 3' end sequence of the nitrilase gene.
[0051] Design three specific primers P5, P6 and P7 according to the Nit M sequence obtained above, use the random primers used in the above three primers and the Tail-PCR method, to Pseudomonas putida ( Pseudomonas putida ) XY4 (preservation number CGMCC No.3830) total DNA as a template to clone the Nit 3' end sequence:
[0052] P5: 5'-CGATGGCGGCAGCCTCTACATGAG-3'
[0053] P6: 5’- GACGAAAGCTGAAACCCACTCATGTA-3’
[0054] P7: 5'-CCAGACCCCAACCAAATACGCGATGT-3'
[0055] The PCR reaction was carried out in a 50 μL system, and the reaction conditions were those used in the Tail-PCR method. The first round of PCR was carried out with P5 and non-specific primers AP1, AP2, AP3 and AP4 respectively. The PCR products amplified by P5 and AP2 were selected as templates, and P6 and NAP were used as primers for the second round of amplification. The third round of PCR was perfo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com