Molecular marking method capable of simultaneously predicting and identifying fineness and crimpness of sheep wool
A technology of molecular markers and curly degree, applied in the field of animal molecular genetics, to achieve the effects of low cost, accelerated breeding process, and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0019] Embodiment 1: In this embodiment, a molecular marker method capable of simultaneously predicting and identifying the fineness and crimp of sheep wool is carried out according to the following steps:
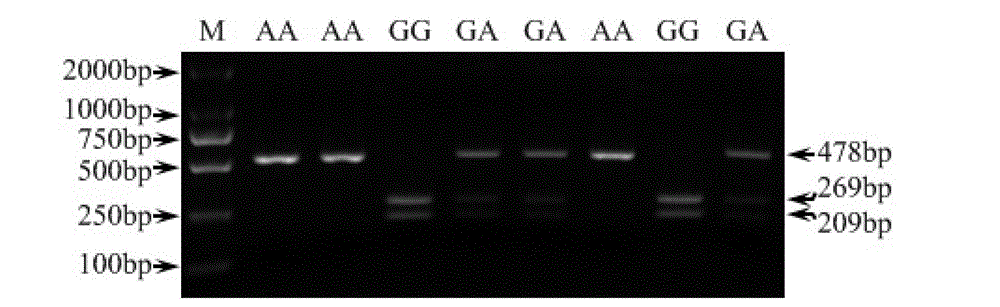
[0020] 1. Design a pair of primers MTRF1 and MTRR1 according to the G9542341A site of the 27th intron region of the sheep MTR gene, and then perform PCR amplification on the sheep genomic DNA to obtain the PCR amplification product, and then use the endonuclease MspI to digest the PCR amplification product, to obtain enzyme cleavage product;
[0021] 2. Use agarose gel with a concentration of 2% to 3% to electrophoresis separate the digested products, and then determine the genotype according to the electrophoresis separation results. The criteria for determination: ① Electrophoresis presents two bands, the size of which is 269bp and 209bp, When the G9542341A site in the 27th intron region of the sheep MTR gene is not mutated, the PCR amplification product can be completel...
specific Embodiment approach 2
[0031] Specific embodiment two: the difference between this embodiment and specific embodiment one is that the reaction system for PCR amplification in step one is a 10 μL reaction system, which consists of the following components:
[0032]
[0033]
[0034] PCR amplification conditions were: 94°C pre-denaturation for 5 min, 94°C denaturation for 30 s, 65°C annealing for 25 s, 72°C extension for 30 s, a total of 33 cycles, 72°C extension for 7 min, and 4°C incubation. Others are the same as in the first embodiment.
specific Embodiment approach 3
[0035] Specific embodiment three: the difference between this embodiment and specific embodiment one is that the enzyme digestion system in step one is as follows:
[0036]
[0037] The enzyme digestion conditions are: 37°C enzyme digestion for 1-2h or overnight. Others are the same as in the first embodiment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com