Related serum microribonucleic acid marker for human severe preeclampsia and application of marker
A technology of preeclampsia and markers, applied in the fields of genetic engineering and clinical medicine, can solve problems that have not received corresponding attention
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Embodiment 1 Research Object Selection and Grouping Basis
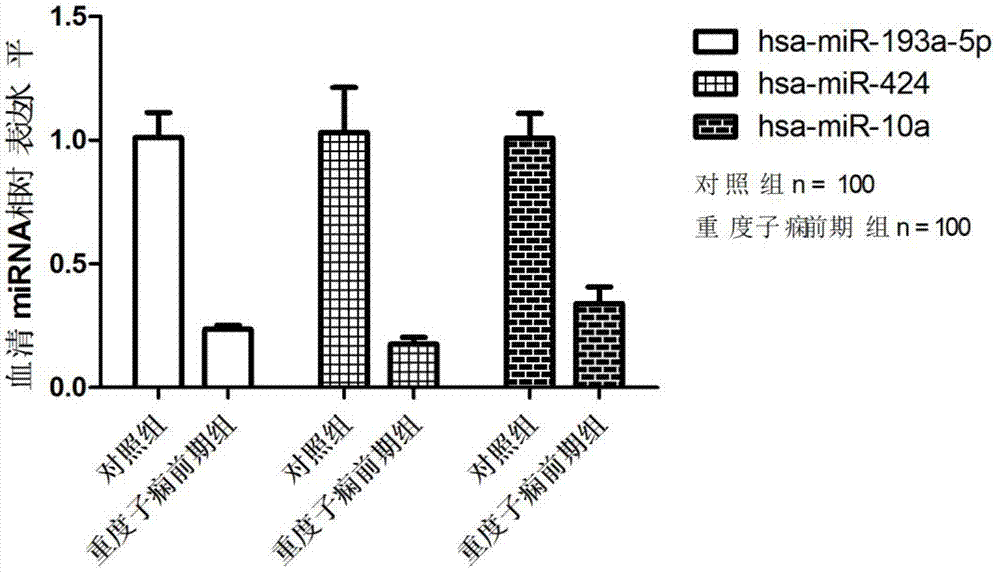
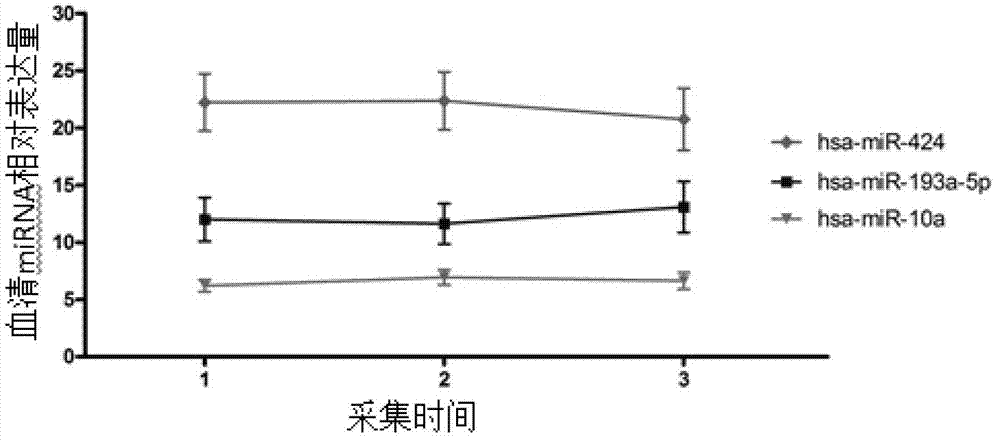
[0077] From July 2009 to October 2011, the inventor collected the blood samples of severe preeclampsia patients and healthy control pregnant women who met the requirements from the Affiliated Hospital of Nanjing Medical University three times. 100 healthy controls (average age: 27.63±3.25 days) and 100 patients with severe preeclampsia (average age: 28.56±4.15 days) were used as experimental subjects for Real-time PCR detection of miRNA expression. Specific sample classification criteria are as follows:
[0078] Group A: healthy control group (n=100, 20 people for microarray screening, 40 people for first-stage verification, 40 people for independent population verification):
[0079] 1. No other major systemic diseases.
[0080]Group B: Severe preeclampsia group (n=100, 20 people for microarray screening, 40 people for phase one verification, 40 people for independent population verification):
[0081] 1. C...
Embodiment 2
[0083] Example 2 TaqMan miRNA array screening
[0084] Preparation of cDNA samples: a) Take 100 μl of serum; b) Add 900 μl of Trizol, shake and mix, centrifuge at 12,000 rpm at 4°C for 15 minutes, and discard the waste liquid in the lower layer; c) Add 1.5 times the volume of supernatant, shake and mix, transfer To the spin column, centrifuge at 12000rpm for 15 seconds, discard the lower waste liquid; d) add 700μl RWT buffer to the spin column, centrifuge at 10000rpm for 15 seconds, discard the lower waste liquid. e) Add 500 μl of RPE buffer to the spin column, centrifuge at 10,000 rpm for 15 seconds, and discard the lower layer. f) Repeat e. g) Add the spin column to a new 2ml tube and centrifuge at 10,000rpm for 1 minute to remove the RPE buffer. h) Add 50 μl of DEPC-treated water to the column and collect RNA by centrifugation at 12000 rpm. i) Then cDNA is obtained by RNA reverse transcription reaction. The reverse transcription reaction system included 4 μl 5×AMV buffe...
Embodiment 3
[0095] Embodiment 3Real-time PCR method measures serum miRNA expression
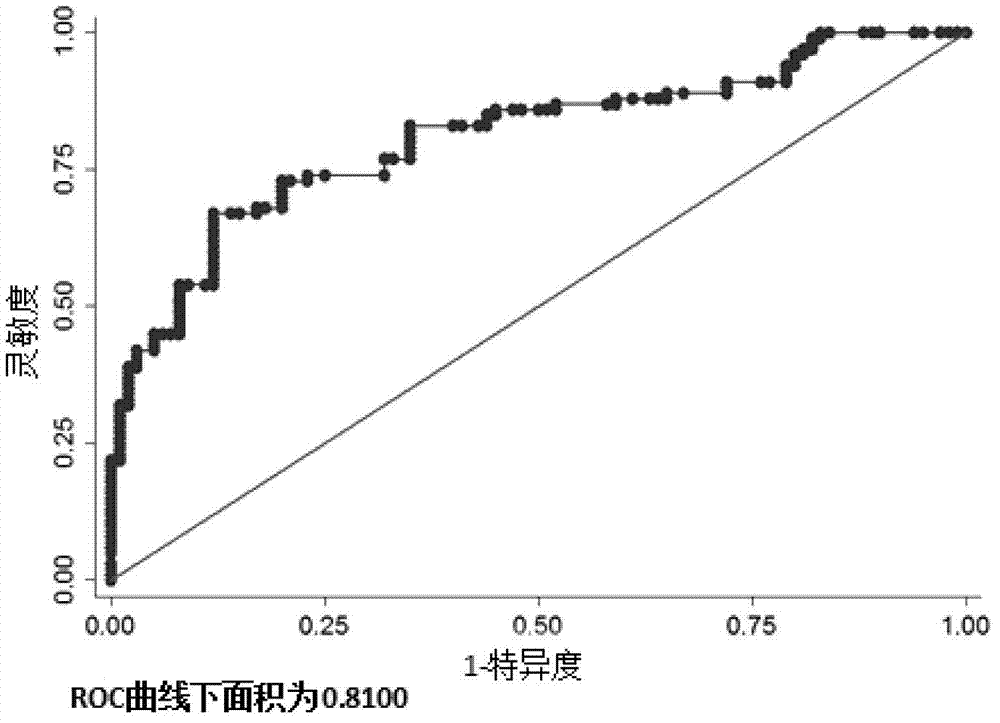
[0096] Primers (Table 1) were designed for quantitative Real-time PCR detection of each miRNAs in the serum of 80 healthy controls and 80 patients with severe preeclampsia.
[0097] (1) Preparation of cDNA samples: a) Take 100 μl of serum; b) Add 900 μl of Trizol, shake and mix, centrifuge at 12,000 rpm at 4°C for 15 minutes, and discard the waste liquid in the lower layer; c) Add 1.5 times the volume of supernatant in absolute ethanol, shake and mix Mix well, transfer to the spin column, centrifuge at 12000rpm for 15 seconds, discard the lower waste liquid; d) add 700μl RWT buffer to the spin column, centrifuge at 10000rpm for 15 seconds, discard the lower waste liquid. e) Add 500 μl of RPE buffer to the spin column, centrifuge at 10,000 rpm for 15 seconds, and discard the lower layer. f) Repeat e. g) Add the spin column to a new 2ml tube and centrifuge at 10,000rpm for 1 minute to remove the RPE buff...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com