Serratia marcescens ankyrin repeat and its application
A technology of Serratia marcescens and ankyrin, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Preparation of Serratia marcescens ankyrin repeat
[0027] Take out the Serratia marcescens PL-06 preserved in the refrigerator at 4°C, pick a single clone and inoculate it into LB liquid medium, culture it overnight at 37°C, 200r / min for 10-12 hours, observe under microscope and identify 16SrDNA, confirm The homology with the 16SrDNA of various Serratia marcescens published in GenBank is 99%, thus confirming that the strain is Serratia marcescens, the microscopic picture is as follows Figure 5 , the 16SrDNA sequence is as sequence 1 in the sequence listing, genbank accession number: JX138534.
[0028] According to the published Serratia marcescens ankyrin repeat subsequence (GenBank: AY091641.1) of the complete genome sequence, the upstream primer SF and the downstream primer SR were designed as reference:
[0029] Upstream primer SF: 5'- CATGCCATGGTA CCTGAAGGGCGTCGCTT -3'
[0030] Downstream primer SR: 5'- CCCCTCGAGA CTGCTGCGCGTAGTG -3'.
[0031] ...
Embodiment 2
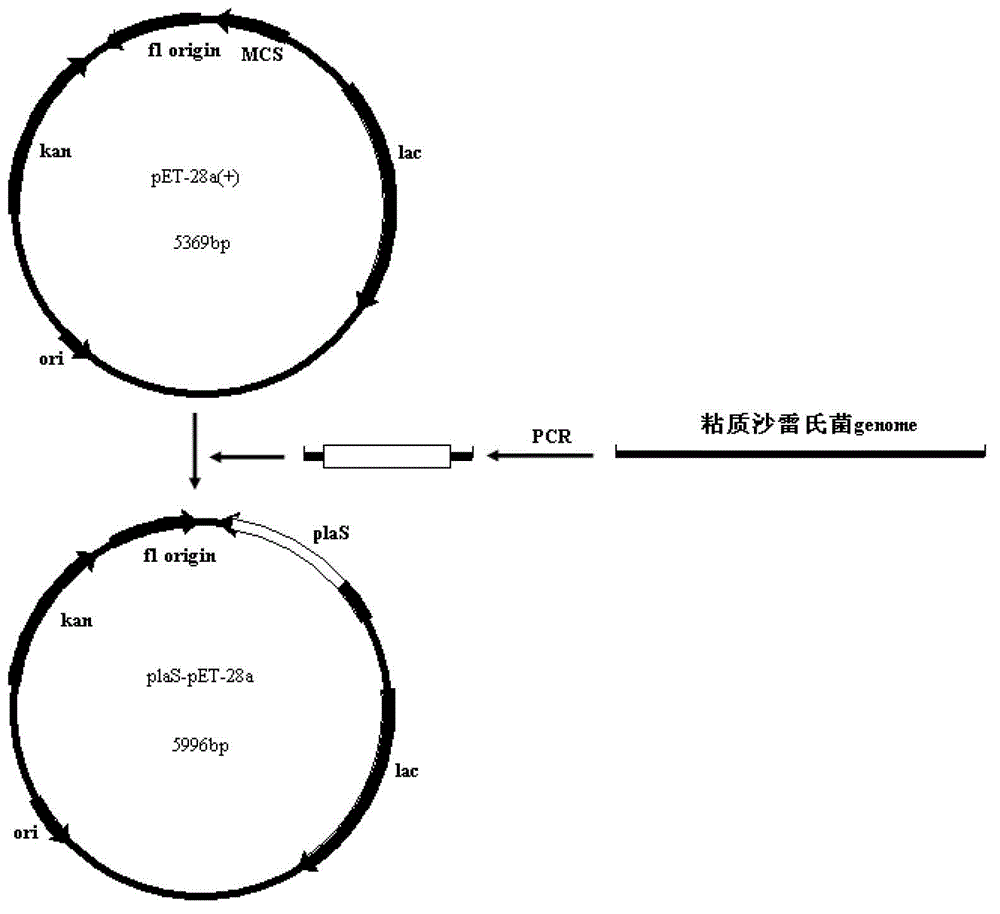
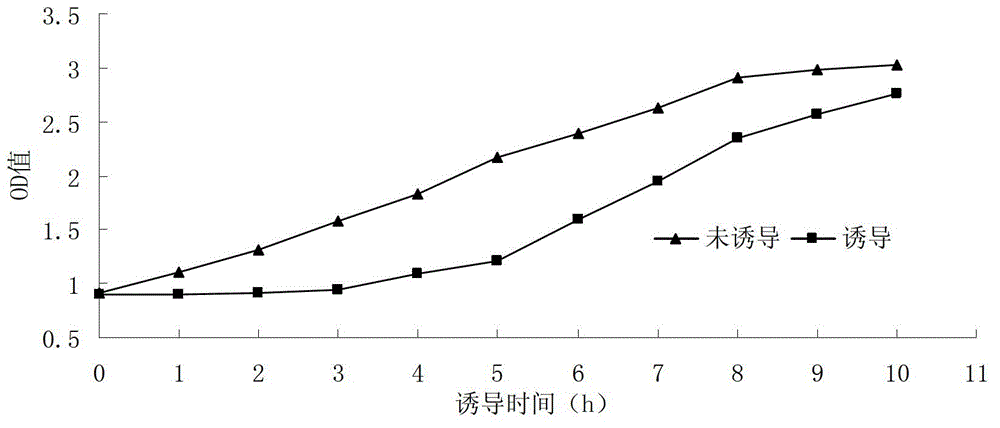
[0044] The construction of embodiment 2 recombinant bacterial strain plaS-pET-28 / BL21 and its thalline growth situation
[0045] ① The product amplified in Example 1 Pla Gel recovery with restriction endonuclease Nco I. xho I double enzyme digestion of the above recovered product and pET-28a (+) plasmid vector, the enzyme digestion system is as follows:
[0046] DNA 10μL
[0047] Nco I 1 μL
[0048] xho I 1 μL
[0049] Buffer K 5μL
[0050] Sterilized double distilled water 33μL
[0051] Total volume 50 μL.
[0052] After mixing, incubate at 37°C for 2 hours. After the end, add 10 μL 6×Loading Buffer to each 50 μL reaction solution to terminate the reaction, recover the digested fragments (about 0.7 kb and 5.4 kb), and connect the two fragments with T4 DNA ligase. The reaction system is as follows:
[0053] PCR double digestion product 12μL
[0054] pET-28a (+) double digestion product 8μL
[0055]Ligase 2μL
[0056] 10×Ligase Buffer 5μL
[0057] Steriliz...
Embodiment 3
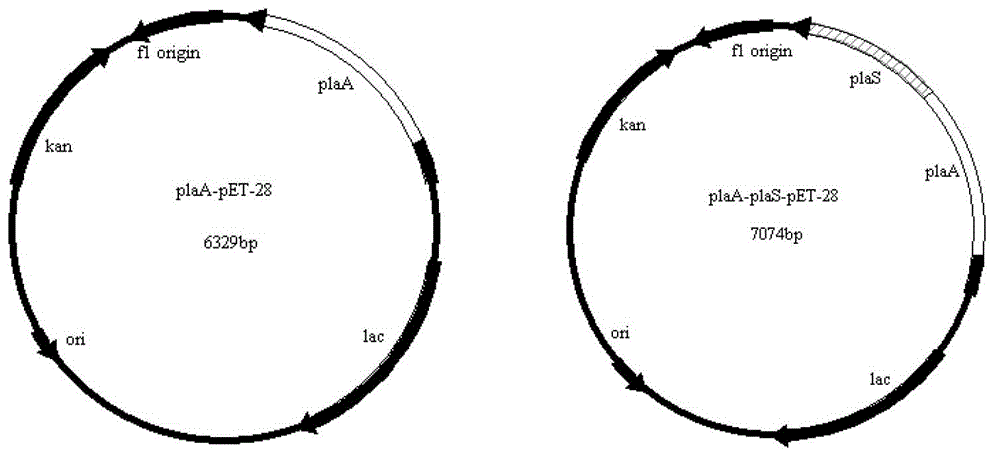
[0062] Embodiment 3 Construction of recombinant bacterial strains plaA-pET-28 / BL21 and plaA-plaS-pET-28 / BL21 and their cell growth
[0063] ① Using the above-mentioned Serratia marcescens PL-06 genome as a template, PCR amplification was performed to obtain the phospholipase A1 gene plaA (see sequence 4 in the sequence listing) and phospholipase A1 gene + ankyrin repeat subgene i.e. plaA-plaS (See Sequence 5 in the Sequence Listing), wherein the underlined part in Sequence 4 is plaA sequence and Pla The part of sequence cross-common, the part underlined in sequence 5 is Pla sequence;
[0064] Among them, PCR amplification plaA Primers when:
[0065] Upstream primer AF: CATGCCATGGGCAGTATGCCTTTAAGT,
[0066] Downstream primer AR: CCCCTCGAGAGGCATTGGCCTTCGCCTC.
[0067] The amplification conditions are as follows:
[0068] AF 1μL
[0069] AR 1 μL
[0070] 10× PCR Buffer 5μL
[0071] 10mmol / L dNTP 2μL
[0072] 25mmol / L MgCl 2 4μL
[0073] Taq DNA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com