CeDGAT1 (Chlorella ellipsoidea Diacylglycerol Acyltransferase 1) gene and application thereof
A kind of gene and amino acid technology, applied in CeDGAT1 gene and its application field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1. Obtaining the full-length cDNA of Chlorella ellipsoide CeDGAT1 gene
[0027] Take the algae liquid of Chlorella ellipsoides (from the Institute of Hydrobiology, Chinese Academy of Sciences) in logarithmic growth phase that has been cultured for 3-4 days, collect the algae bodies by centrifugation, and place them in liquid nitrogen to grind them thoroughly. Total RNA was extracted and purified according to the instructions of Aidelai EASYspin RNA Extraction Kit and the instructions of Takara DNase I. cDNA was obtained by reverse transcription with polyT primers, and RT-PCR was performed (see TOYOBO ReverTra Ace-α Kit for specific operations).
[0028] The cDNA synthesis reaction conditions are as follows:
[0029]
[0030] RNase Free H 2 O to a total volume of 20 μL. After flicking to mix well and centrifuging briefly, perform the reverse transcription reaction as follows:
[0031]
[0032] An appropriate amount of the above reverse transcription pro...
Embodiment 2
[0036] Example 2. Construction of Yeast Expression Vector Containing CeDGAT1 Gene
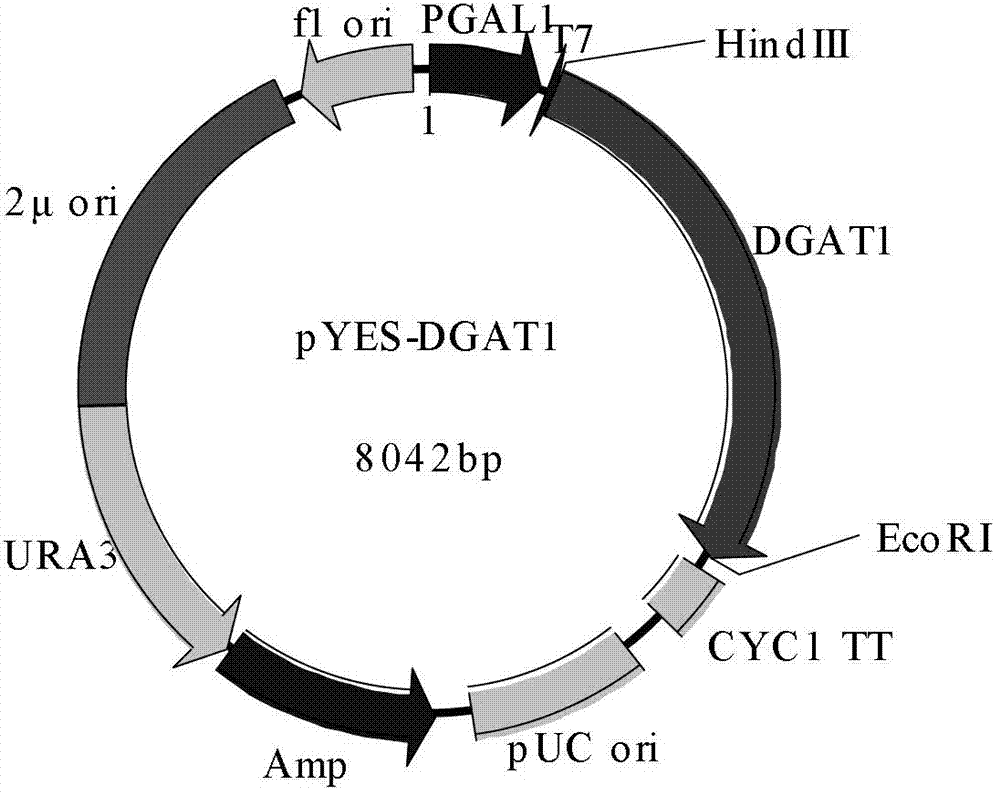
[0037] According to the cds sequence of CeDGAT1, primers with Hind III and EcoR I restriction sites were designed (the sequences of the two primers are: cgccAAGCTTATGCCAGATGATGCCAGCA and ccggGAATTCTCAGCTGCCATTTGCGAG, respectively). From the pEASY-Blunt vector containing CeDGAT1, high-fidelity EasyPfu DNA Polymerase (purchased from Transgen Company) was used to amplify the fragment containing Hind III and EcoR I restriction sites, and Hind III and EcoR I (purchased from Takara Company) were used to amplify the fragment. ) was subjected to double enzyme digestion, connected with the yeast expression vector pYES2 (purchased from Invitrogen Company) of the same double enzyme digestion, sequenced and verified, and named as pYES-DGAT1, the vector diagram is shown in figure 1 .
Embodiment 3
[0038] Example 3. Transformation of Saccharomyces cerevisiae with yeast expression vector pYES-DGAT1
[0039] Inoculate uracil-deficient Saccharomyces cerevisiae INVSC1 (purchased from Invitrogen) into 10 mL of YPD medium, and culture overnight at 30° C. with shaking. Inoculate the bacterial solution into 50mL YPD medium the next day and dilute to OD 600 =0.4, continue to culture for 2-4h to OD 600 Between 0.5-0.6, 5,000rpm refrigerated centrifugation for 1min, suspended the pellet with 40mL 1×TE (10mM Tris, pH7.5, 1mM EDTA), 5,000rpm refrigerated centrifugation, and 2mL 1×LiAc (10mM lithium acetate, pH7.5) / Suspend the precipitate with 0.5×TE and place it at room temperature for 10 minutes. Mix 100 μL of yeast suspension with 1 μg of yeast expression vector pYES-DGAT1 and 100 μg of denatured salmon sperm DNA, then add 700 μL of 1×LiAc / 40% PEG-3350 / 1×TE, and mix well. Incubate at 30°C for 30 minutes, add 88 μL DMSO, mix well, and heat shock at 42°C for 7 minutes. Centrifug...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com