Novel anti-HBV endogenous protein HNF1alpha and application thereof
A protein and protein technology, applied in the direction of application, peptide/protein composition, peptide source, etc., can solve the problem of not finding hepatitis B
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
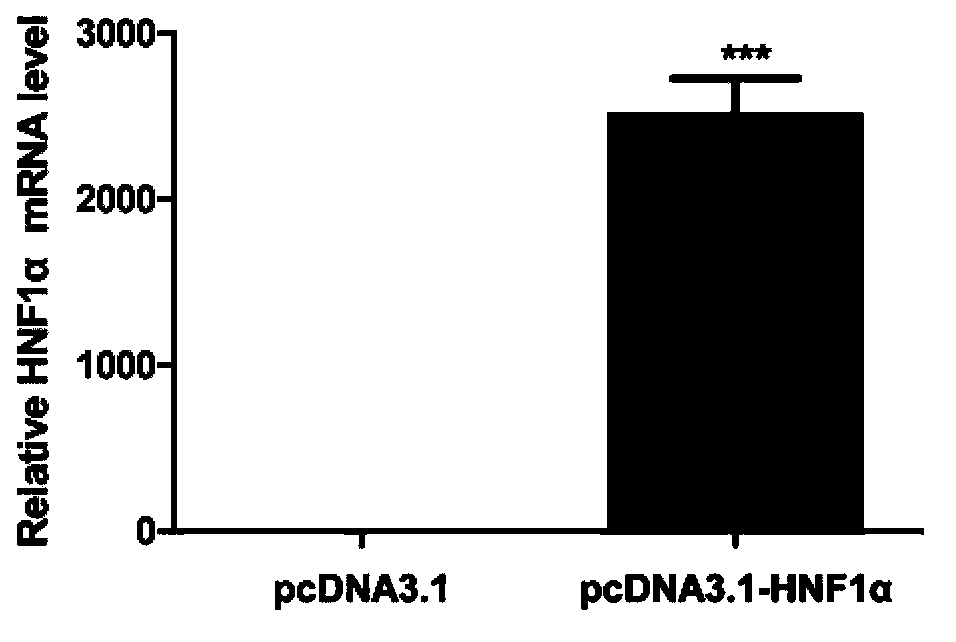
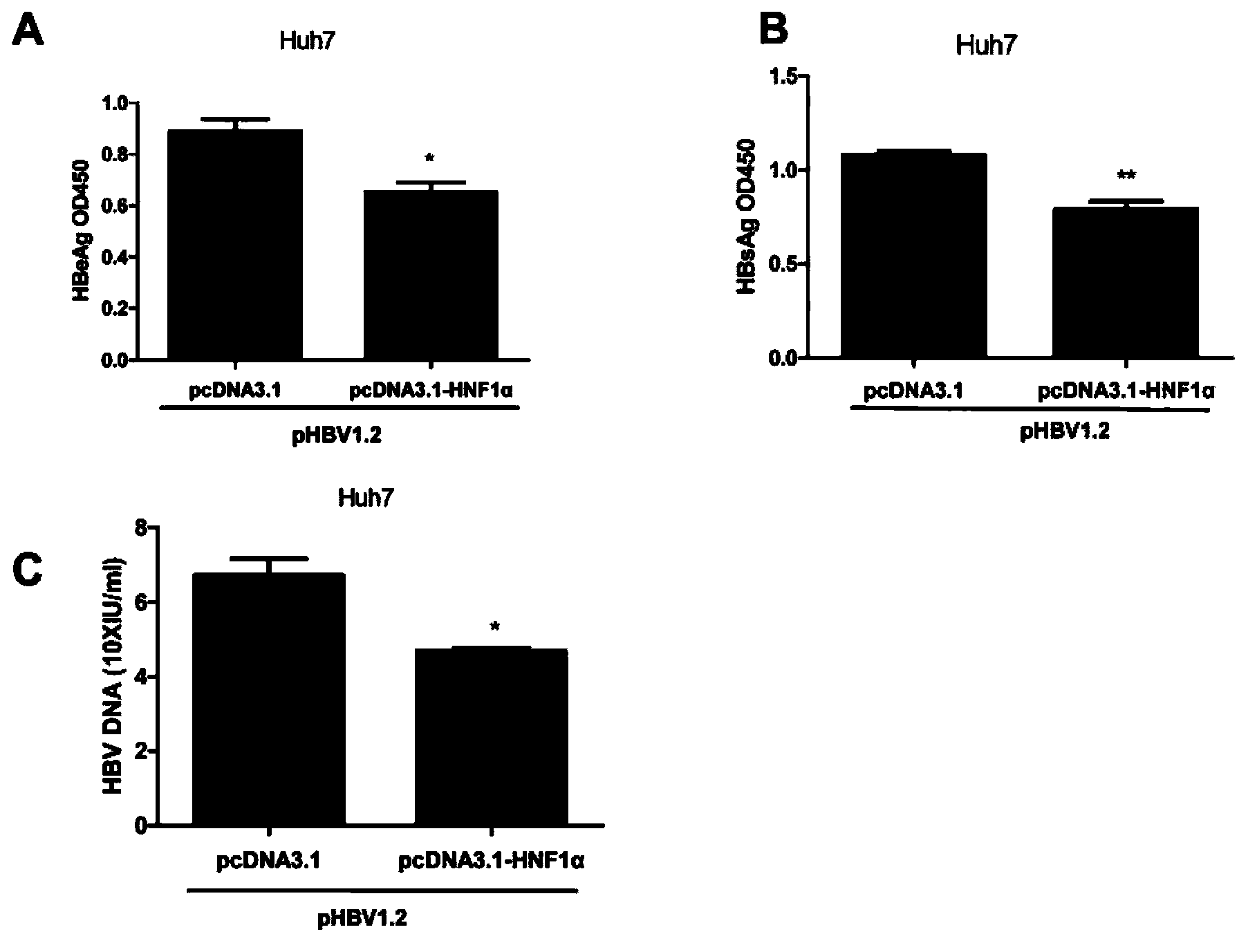
[0036] Example 1. Overexpression of HNF1α to detect the effect of HNF1α on HBV replication
[0037] 1. Construction of HNF1α eukaryotic expression vector
[0038] (1) Total RNA of Huh7 was extracted by Trizol method and reverse transcribed into cDNA.
[0039] (2) Using cDNA as a template, the HNF1α gene was amplified with primers HNF1αP1 and HNF1αP2 (see Table 1).
[0040] The amino acid sequence of HNF1α is shown in SEQ ID No.1, and the amplified HNF1α gene sequence is shown in SEQ ID No.2.
[0041] PCR program: 95°C 5min; (95°C 30s, 60°C 30s, 72°C 2min) 30cycles; 72°C 7min.
[0042] (3) Perform 1% agarose gel electrophoresis on the PCR product, and after the correct size is identified, cut the gel and recover.
[0043] (4) Ligate the recovered product with the pMD18-T vector to obtain a recombinant plasmid, and transform the recombinant plasmid into Escherichia coli DH5α.
[0044] (5) Screen the positive clones with the ampicillin plate, pick a single clone, extract the ...
Embodiment 2
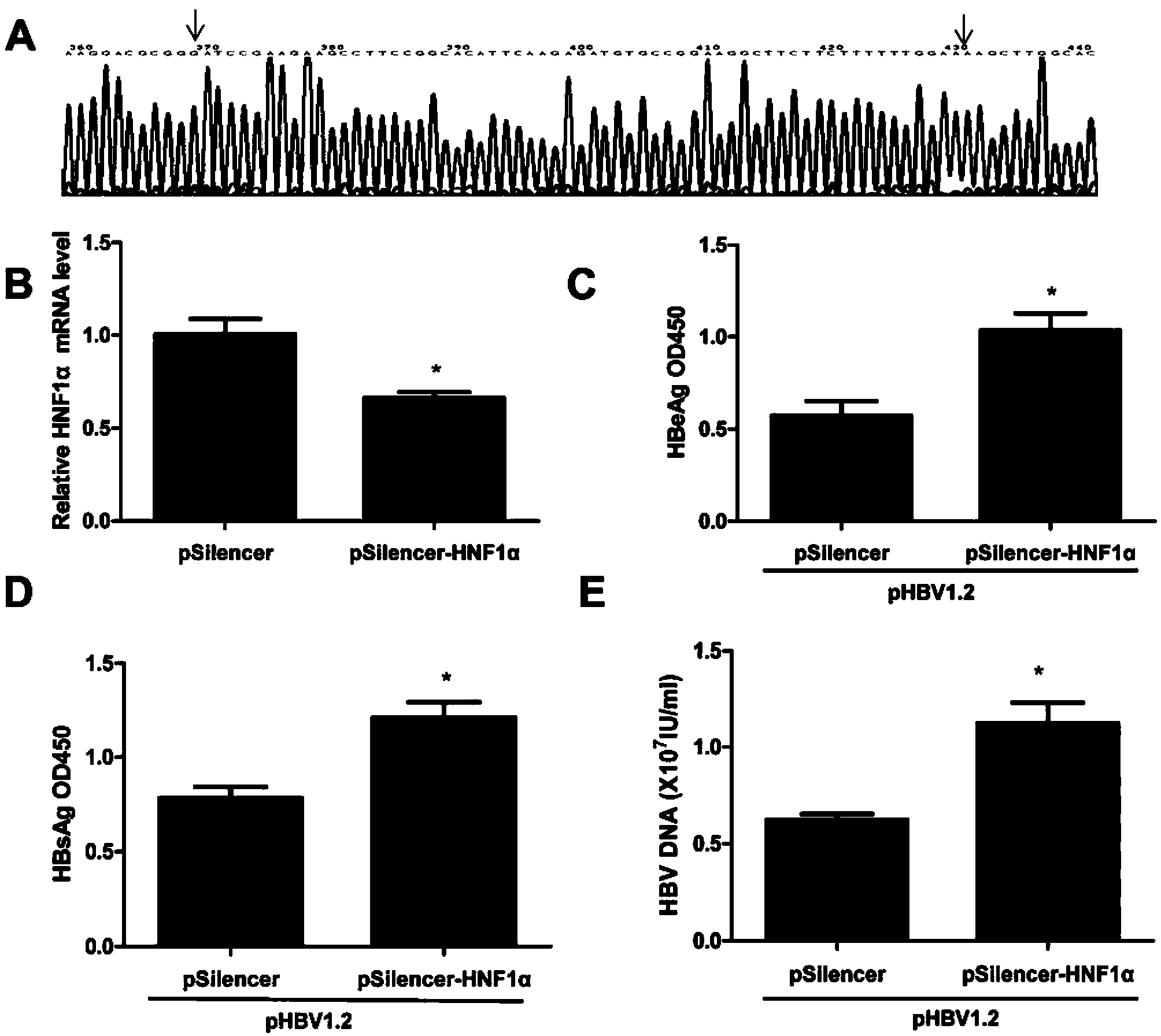
[0072] Example 2. Inhibition of the expression of HNF1α and detection of its effect on HBV replication
[0073] 1. Construction of pSilencer-HNF1α
[0074] (1) Synthesize the primers siHNF1αP1 and siHNF1αP2 (see Table 1) of the HNF1α inhibitory vector, and anneal the primers.
[0075] (2) Digest the pSilencer2.1-U6hygro (hereinafter referred to as pSilencer) vector with BamHI and HindIII to obtain a large fragment of the vector, and connect the annealed primers and the large fragment of the vector to obtain a recombinant plasmid.
[0076] (3) The recombinant plasmid was transformed into Escherichia coli DH5α, positive clones were screened with ampicillin, the plasmid was extracted after 12 hours of liquid culture, and the sequence was determined after identification by BamHI and HindIII digestion, and the sequenced vector was named pSilencer-HNF1α.
[0077] The sequencing results of the inhibitory sequence in pSilencer-HNF1α are as follows: image 3 As shown in A, pSilencer-...
Embodiment 3
[0098] Example 3, Interaction Analysis of HNF1α and Enhancer I (EnhancerI, EnhI) of HBV
[0099] 1. Construction of pGL3-EnhI
[0100] (1) The six regulatory elements S1, S2, CORE, X promoter, enhancer I and enhancer II of HBV were analyzed using MatInspector. The analysis results showed that enhancer I (including the X promoter region of HBV) contained HNF1α binding site.
[0101] (2) Using pHBV1.2 as a template, using EnhancerI P1 and EnhancerI P2 as primers (see Table 1 for the sequence) to amplify HBV enhancer I, perform 1% agarose electrophoresis on the amplified product, and recover a band of about 411bp from the gel . The sequence of the amplified enhancer I is shown in SEQ ID No.3.
[0102] (3) The PCR amplification product was connected to the pMD18-T vector to obtain the recombinant plasmid pMD18-T-EnhI. The recombinant plasmid was transformed into Escherichia coli DH5α, positive clones were screened with ampicillin plate, the plasmid was extracted after 12 hours...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com