A detection method for surface-enhanced Raman spectroscopy of low-abundance proteins in plasma
A technology for surface-enhanced Raman and low-abundance proteins, which is applied in the field of low-abundance proteins, can solve the problems of expensive mass spectrometers, inability to identify and distinguish proteins of equal quality, and complicated steps, etc., to make up for the identification of low-abundance proteins, rich The collection process is simple and easy, and the effect of improving detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
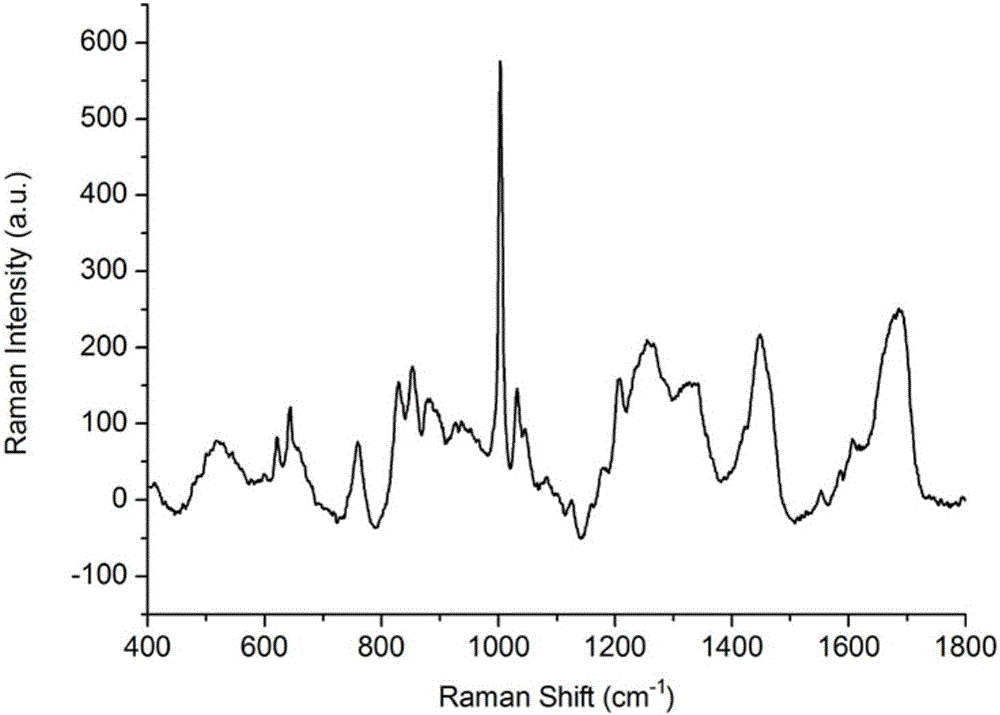
[0037] 1. SERS spectrum detection of low-abundance proteins in plasma of liver cancer patients based on silver nanoparticles enhanced substrate
[0038] (1) Removal of high-abundance proteins in the plasma of patients with liver cancer
[0039]Take 2 g of ion exchange resin in a collection tube covered with a spin column, add 100 μl of eluent, mix well, centrifuge at 3000 rpm for 5 min, pour out the liquid in the collection tube, and repeat three times. Take 10 μl of liver cancer patient plasma standard sample and dilute it in 100 μl of eluent, mix it evenly, add it to the spin column containing ion exchange resin, use a pipette to repeatedly suck and mix well, place it on a shaker at 200rpm for 15min, and shake it at 2000rpm The plasma low-abundance protein solution of patients with liver cancer was collected by centrifugation.
[0040] (2) Enrichment of low-abundance proteins in plasma of patients with liver cancer
[0041] Take 0.2 g of cellulose acetate membrane and incu...
Embodiment 2
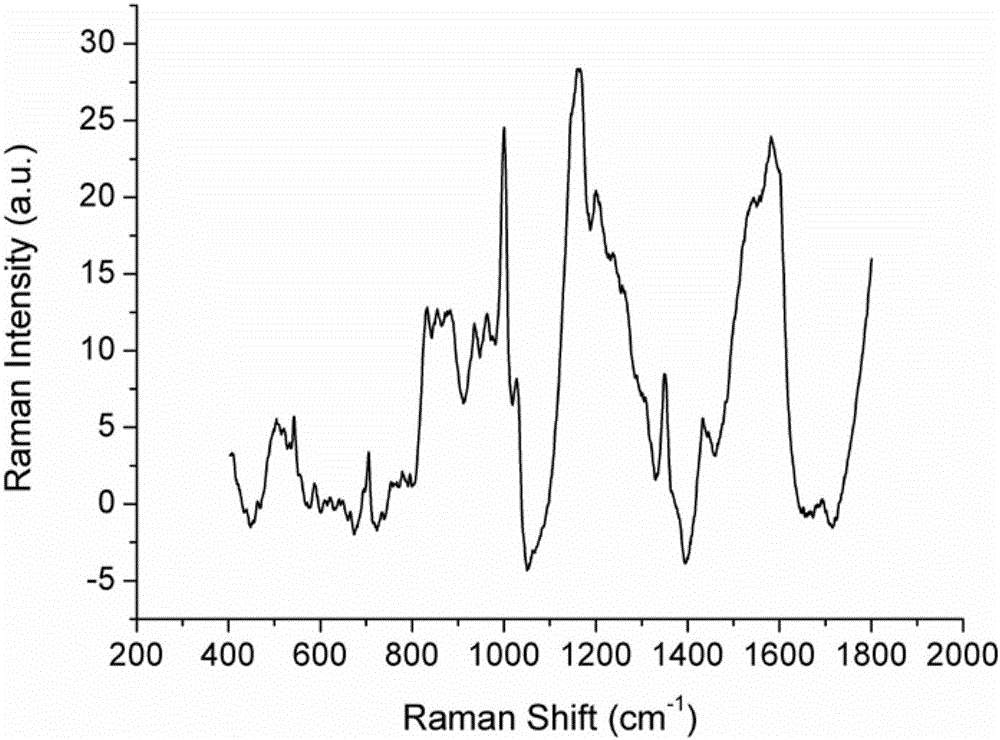
[0047] SERS Spectral Detection of Low Abundance Proteins in Plasma of Nasopharyngeal Carcinoma Patients Based on Gold Nanoparticles Enhanced Substrate
[0048] (1) Removal of high-abundance proteins in the plasma of patients with nasopharyngeal carcinoma
[0049] Take 25g of ion exchange resin in a collection tube covered with a spin column, add 1000μl of eluent and mix well with the hydrophobic polymer, centrifuge at 6000rpm for 1min, pour out the liquid in the collection tube, and repeat three times. Take 100 μl of nasopharyngeal carcinoma patient plasma standard sample and dilute it in 1000 μl of eluent, mix it evenly, add it to the spin column containing ion exchange resin, use a pipette to repeatedly pump and mix well, place it on a shaker at 350rpm for 10min , centrifuge at 5000rpm to collect the plasma low-abundance protein solution of patients with nasopharyngeal carcinoma.
[0050] (2) Enrichment of low-abundance proteins in plasma of patients with nasopharyngeal car...
Embodiment 3
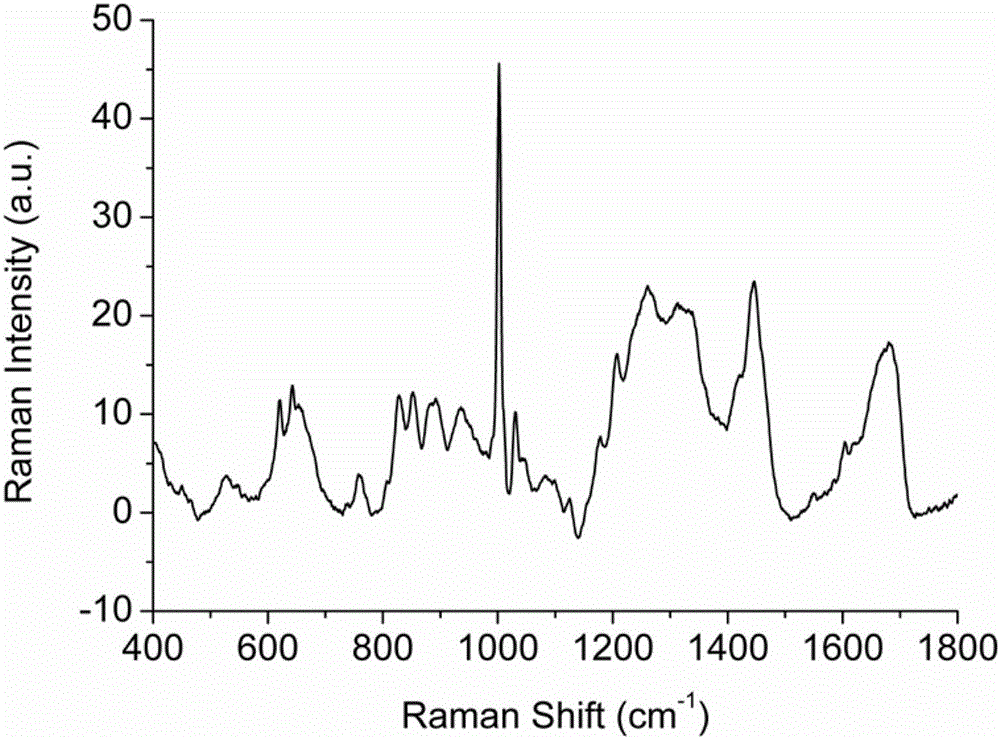
[0057] 1. SERS spectrum detection of low-abundance proteins in plasma of gastric cancer patients based on silver nanoparticles enhanced substrate
[0058] (1) Removal of high-abundance proteins in the plasma of patients with gastric cancer
[0059] Take 2 g of silica gel in a collection tube covered with a spin column, add 150 μl of eluent, mix well, centrifuge at 3000 rpm for 5 min, pour out the liquid in the collection tube, and repeat three times. Take 10 μl of gastric cancer patient plasma standard sample and dilute it in 100 μl of eluent, mix it evenly, add it to the spin column containing silica gel, use a pipette to repeatedly suck and mix well, place it on a shaker at 200rpm for 15min, and collect it by centrifugation at 2000rpm Plasma low-abundance protein solution in patients with liver cancer.
[0060] (2) Enrichment of low-abundance proteins in plasma of patients with gastric cancer
[0061] Take 2 g of nitrocellulose membrane and incubate with the plasma low-abu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com