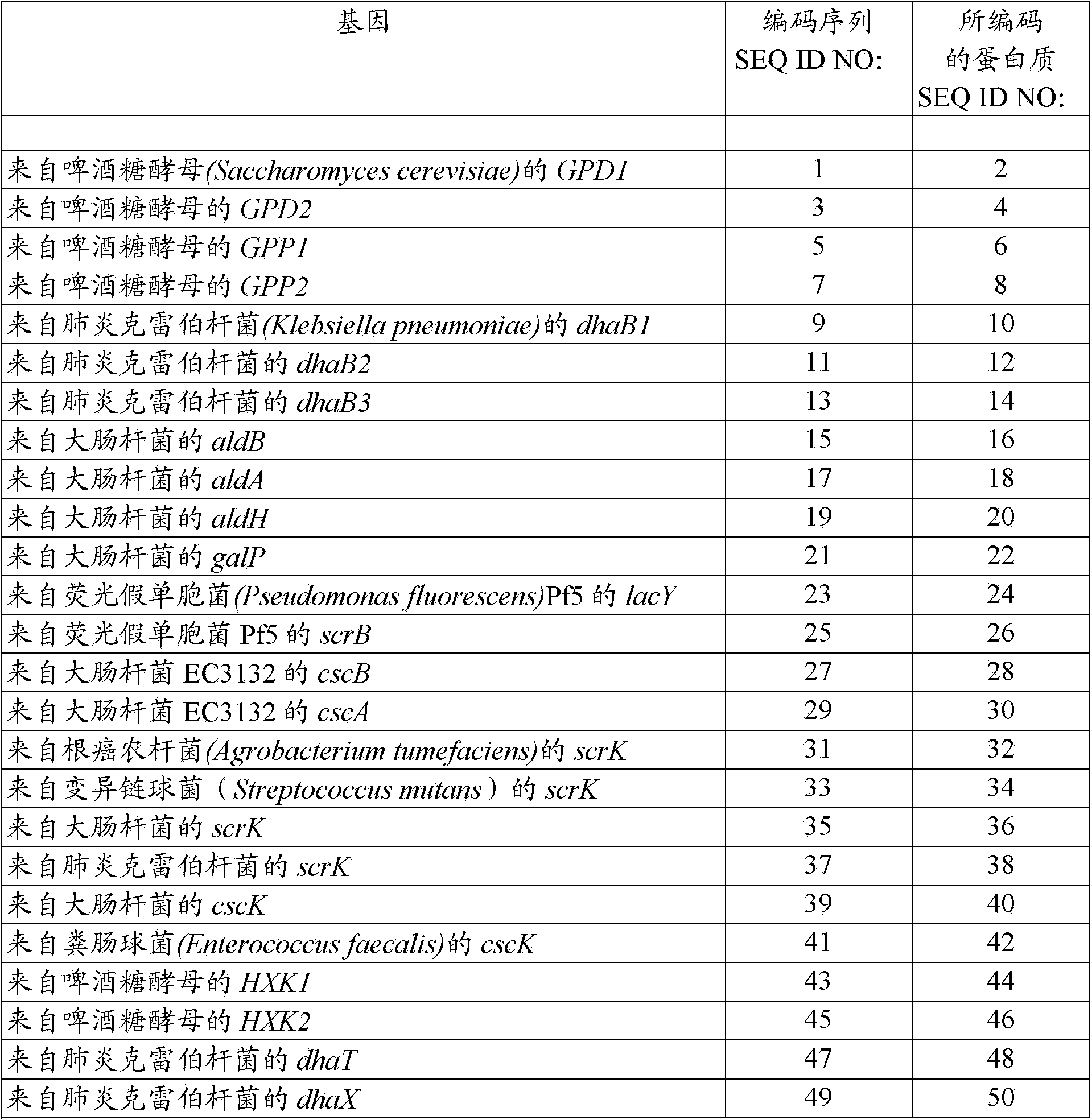
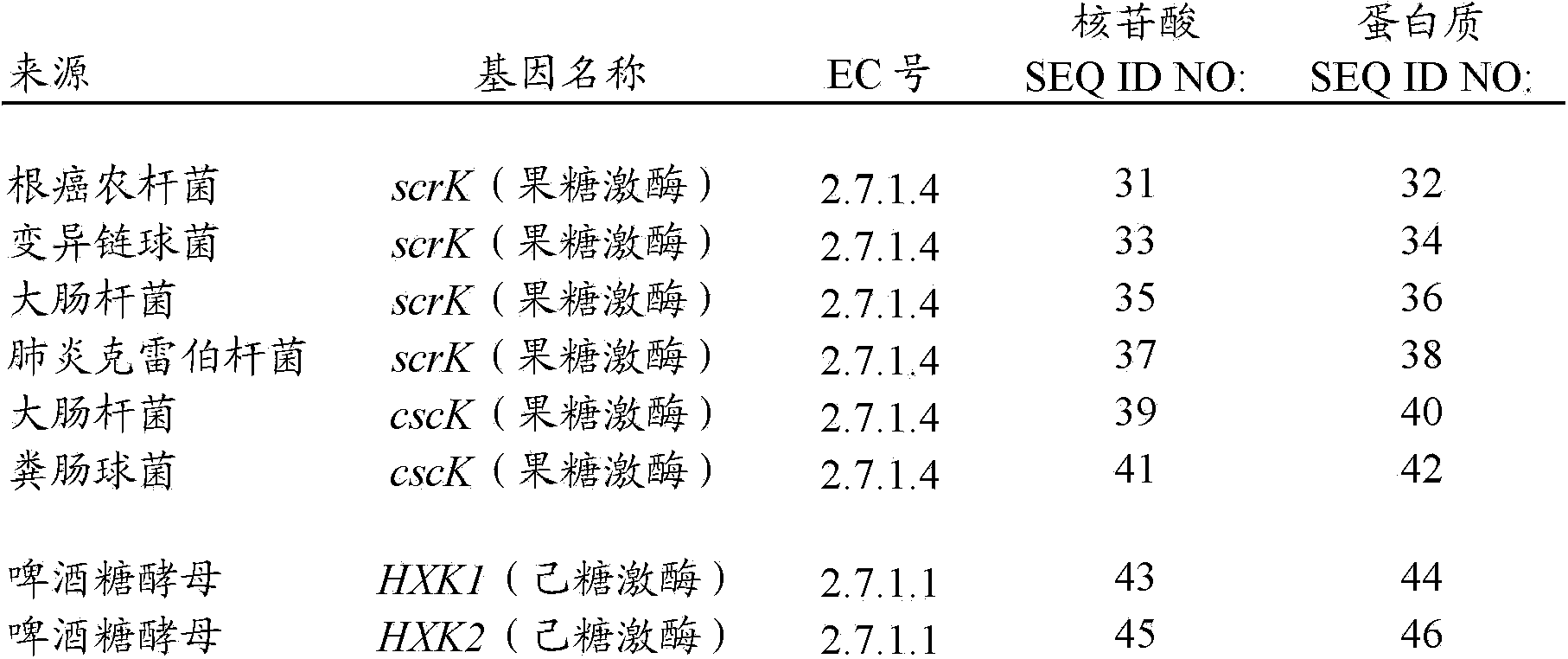
Recombinant bacteria having the ability to metabolize sucrose
A technology for recombining bacteria and sucrose, applied in the fields of microbiology and molecular biology, can solve the problems of lack of utilization of sucrose
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0218] The invention will be further illustrated in the following examples. It should be understood, that these Examples, while indicating preferred embodiments of the invention, are given by way of illustration only. From the above discussion and these Examples, one skilled in the art can ascertain the characteristics of this invention, and without departing from the spirit and scope thereof, can make various changes and modifications of the invention to adapt it to different usage and conditions .
[0219] general method
[0220]Standard recombinant DNA and molecular cloning techniques described in the Examples are well known in the art and are described more fully in: Sambrook, J., Fritsch, E.F. and Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, (1989) (Maniatis), with T.J. Silhavy, M.L. Bennan, and L.W. Enquist, Experiments with Gene Fusions, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. (1984) an...
example 1
[0228] Recombinant Escherichia coli containing putative sucrose metabolism genes
[0229] The purpose of this example is to construct the BAA-477) a putative sucrose metabolism gene and a recombinant E. coli strain capable of producing 1,3-propanediol (PDO) from sucrose.
[0230] Using the high-fidelity polymerase Phusion TM Flash (Finnzymes Oy, Espoo, Finland), the Pseudomonas fluorescens Pf5 genes lacY (SEQ ID NO: 23) and scrB (SEQ ID NO: 25) (which are adjacent genes) were obtained by PCR amplification using the following conditions and the promoter region upstream of the Pf5 gene lacY: 98°C / 10 sec; 30 cycles of 98°C / 1 sec, 63°C / 30 sec, and 72°C / 30 sec; then 72°C / 5 min, with fluorescence from The genomic DNA of Pseudomonas Pf5 was used as a template, and the primers used were shown in SEQ ID NO:56 and SEQ ID NO:57.
[0231] One adenosine nucleotide ("A") was added to the 3' end of the 3063 bp PCR product with Taq polymerase at 72°C for 10 minutes. PCR products were ...
example 2
[0239] Production of 1,3-propanediol from sucrose
[0240] The purpose of this example is to demonstrate the presence of BAA-477) an E. coli strain with putative sucrose metabolism genes was able to metabolize sucrose and produce 1,3-propanediol (PDO) and glycerol.
[0241] E. coli strains PDO2241 and PDO2242 described in Example 1 were grown overnight at 33°C in Miller's modified L-Broth (Teknova, Half Moon Bay, CA) supplemented with 100 mg / L spectinomycin and 100 mg / L ampicillin . These cultures were used to inoculate shake flasks at an optical density of 0.01 units measured at 550 nm in MOPS minimal medium (Teknova, Half Moon Bay, CA) supplemented with 11 g / L sucrose. Vitamin B12 was added to the medium to a concentration of 0.1 mg / L. The cultures were incubated at 34°C with shaking (225 rpm) for 24 hours, after which the concentrations of sucrose, glycerol and 1,3-propanediol (PDO) in the above broths were determined by high performance liquid chromatography.
[024...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com