Method for producing bile salt hydrolase by fermentation of twin-arginine signal peptide and application thereof
A bile salt hydrolase, double arginine technology, applied in the directions of hydrolase, biochemical equipment and methods, enzymes, etc., can solve the problems of reducing the expression amount of target protein in fermentation supernatant, and folding can not be carried out effectively.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Embodiment 1: the construction of recombinant bacteria
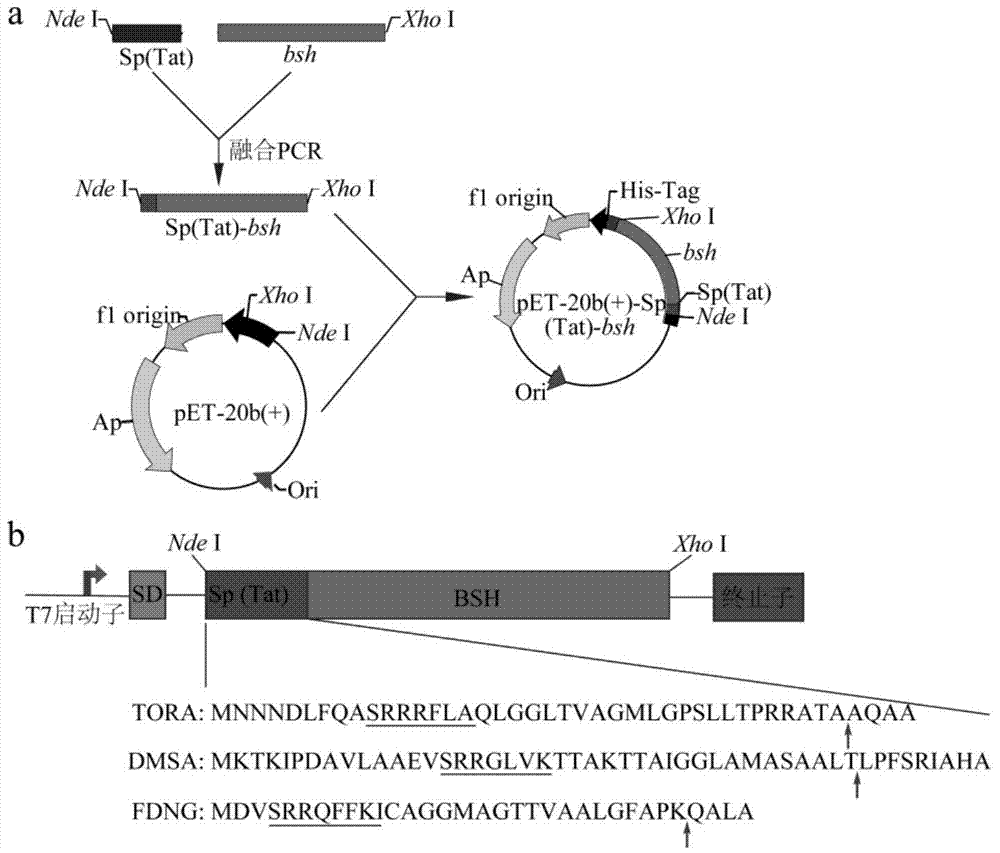
[0075] 1) The construction process of the recombinant plasmid is as follows: figure 1 as shown, figure 1 : (a) Construction process of expression plasmid pET-20b(+)-SP(Tat)-bsh, SP(Tat), signal peptide of Tat pathway; (b) Schematic diagram of fusion protein SP(Tat)-BSH. The signal peptide of the double-arginine pathway is directly fused to BSH. The underlined part is the double arginine motif "SRRXXXX"; the arrow indicates the cleavage site of signal peptidase I.
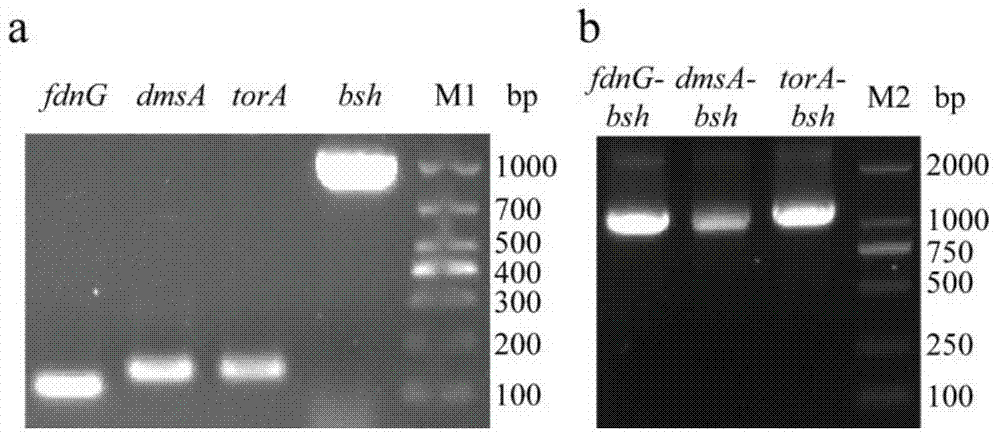
[0076] The specific operation is as follows: using the plasmid pUC57-Tat as a template, using primers torA (F) and torA (R) to amplify the torA gene encoding the TorA signal peptide by PCR ( figure 2 a), PCR conditions are: 95°C pre-denaturation for 10 minutes; 98°C for 10s, 55°C for 30s, 72°C for 20s, 30 cycles; 72°C for 10 minutes. At the same time, using the genomic DNA of L. plantarum BBE7 as a template, using torA-bsh(F) and bsh(R) as primers,...
Embodiment 2
[0079] Example 2: Induced expression of recombinant bacterial BSH and assay of enzyme activity and protein electrophoresis
[0080] The seeds cultivated overnight at 20°C and 200rpm were transferred to 25mL TB medium with a 2% inoculum, and cultivated to OD at 37°C and 200rpm 600 =0.6, add 0.4mM IPTG to induce 6h; through SDS-PAGE and enzyme activity assay, analyze the expression of bile salt hydrolysis enzyme.
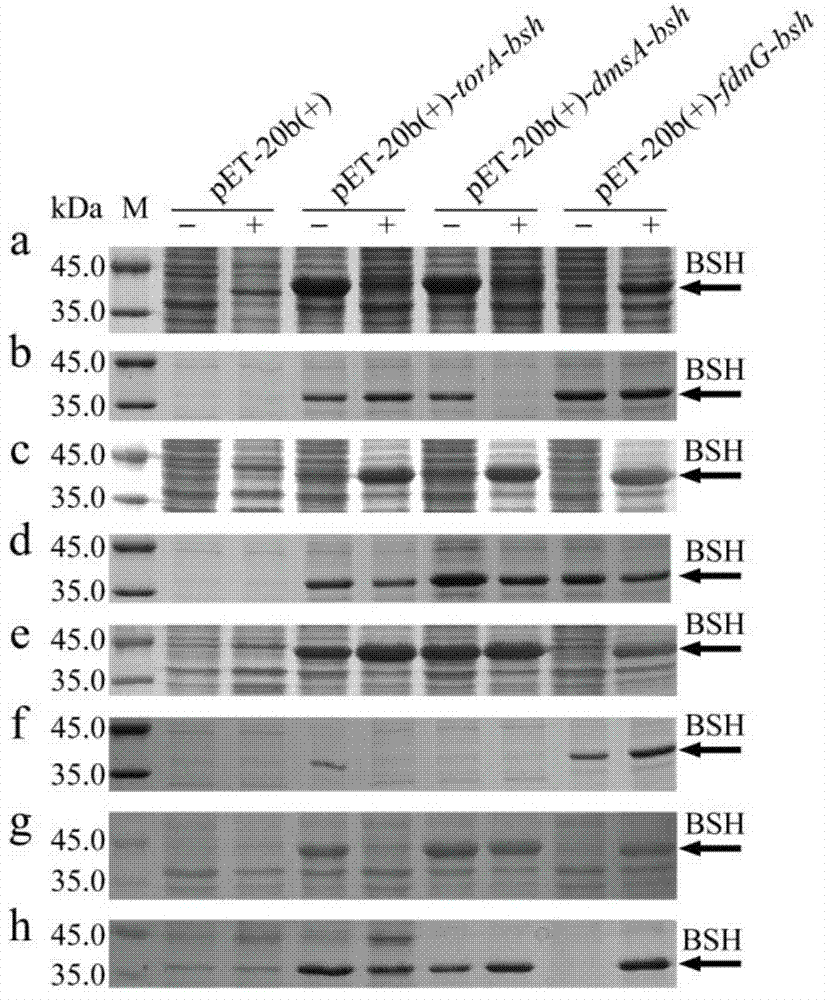
[0081] The effects of three signal peptides on the intracellular expression of BSH were analyzed by SDS-PAGE. The expression of BSH in the recombinant bacterial cell is as follows image 3 a, 3c, 3e and 3g. image 3 (a), (c), (e), (g) are the whole cell protein; (b), (d), (f), (h) are the protein in the fermentation supernatant; M, protein molecular weight standard ; -, not induced with IPTG; +, induced with 0.4 mM IPTG. It can be seen from the figure that BSH is expressed to varying degrees in all recombinant bacteria. However, the molecular weight of these reco...
Embodiment 3
[0091] Embodiment 3: Optimization of BSH activity outside the recombinant bacteria
[0092] In E. coli, the composition of the medium and the fermentation conditions, such as temperature, concentration of inducer, and other parameters, will affect the secretion and expression of heterologous proteins. This chapter uses an orthogonal experimental design (L9(3 4 )) studied the induction temperature, inoculum size, induction OD 600 And the effect of IPTG concentration on the extracellular BSH activity of recombinant E.coli BL21(DE3)pLysS(pET-20b(+)-dmsA-bsh). The experiment was designed and analyzed by the software Orthogonal Design Assistant 3.1.1. The specific experimental design and results are shown in Table 2. These fermentation conditions have a great impact on the activity of recombinant bacterial extracellular BSH, and their influence order is: induction temperature > IPTG concentration > induction OD 600 > Inoculum volume. When induction temperature, IPTG concentrat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com