PCR (Polymerase Chain Reaction) detection primer and method of pseudomonas putida
A technology of Pseudomonas putida, detection methods, applied in the directions of biochemical equipment and methods, determination/inspection of microorganisms, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Primer design:
[0024] Check the complete gene sequence of Pseudomonas putida on Gengbank, and design specific primers for the conserved genes of Pseudomonas putida. The specific nucleotide sequence is:
[0025] Sequence 1: 5'-CTG CAT CAT GGC CGG TGA CAA CAT TT- 3'
[0026] Sequence 2: 5'-GTC GCA TGG CTG TCG GTC TTC AGA TC-3'
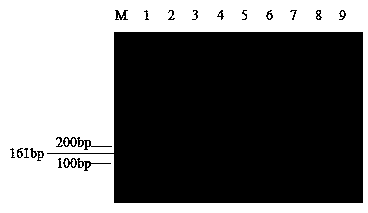
[0027] The amplified target gene fragment of Pseudomonas putida was 161bp.
Embodiment 2
[0029] DNA template extraction of the bacteria liquid to be tested:
[0030] 1) Add 1g (mL) sample into 100mL normal saline and mix evenly by aseptic operation, pipette 1 loop of bacteria and inoculate 5 Pseudomonas chromogenic medium plates continuously, and incubate at 30°C for 24h. During the sampling process, a Pseudomonas chromogenic medium plate was placed next to the sample as a blank control.
[0031] 2) Pick 1-3 single colonies on the plate and transfer them to nutrient broth in a constant temperature incubator at 35±1°C for 18h-24h.
[0032] 3) Extraction of bacterial genomic DNA
[0033] Take 200 ul of the above-mentioned cultured bacterial solution, use a commercial bacterial genomic DNA extraction kit, and refer to the instructions for specific extraction operations to extract bacterial genomic DNA.
Embodiment 3
[0035] Establishment of PCR amplification method:
[0036] 1) Preparation of PCR reaction system: See Table 1 for specific composition preparation.
[0037] Table 1
[0038]
[0039] A negative control and a positive control were also set up.
[0040] 2) PCR reaction conditions:
[0041] Put the above-prepared small tube of PCR reaction solution into the PCR instrument. The reaction conditions are: 94 °C pre-denaturation for 1 min; 94 °C for 15 s, 61 °C for 15 s, 72 °C for 15 s, and 30 cycles; 72 °C for 5 min.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com