A Construction Method of Microsatellite DNA Molecular Marker
A technology of leopard gills and DNA molecules is applied in the field of construction of microsatellite DNA molecular markers for leopard gills, which can solve the problems of limited quantity, time-consuming and labor-intensive, and achieve the effect of short time consumption.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
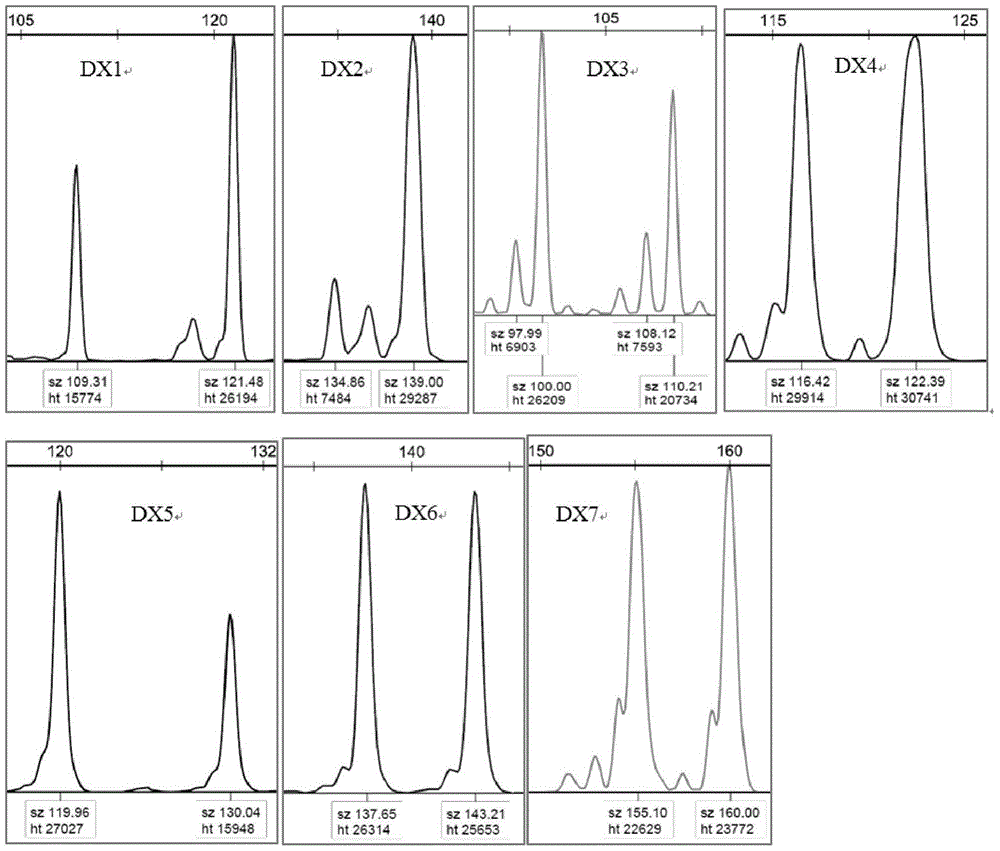
[0022] A method for constructing a microsatellite DNA molecular marker of leopard gill echinacea, comprising the following steps:
[0023] 1. Transcriptome sequencing of leopard gillfish:
[0024] The total RNA of the sample was extracted by the conventional Trizol method, the DNA in the total RNA was digested by DNase I, and the mRNA was enriched by the mutual adsorption between the magnetic beads with Oligo (dT) and the 3' polyA of the mRNA, and the mRNA was interrupted by heating. After the reaction of the reagent, the product was recovered by ethanol precipitation on ice, double-stranded cDNA was synthesized on the PCR machine, and the end was repaired in the Thermomixer to add base A to the 3' end of the repair product, and then add "A" to the adapter and Product ligation, purification, recovery, amplification and library construction, using Illumina Hi Seq TM 2000 sequenced.
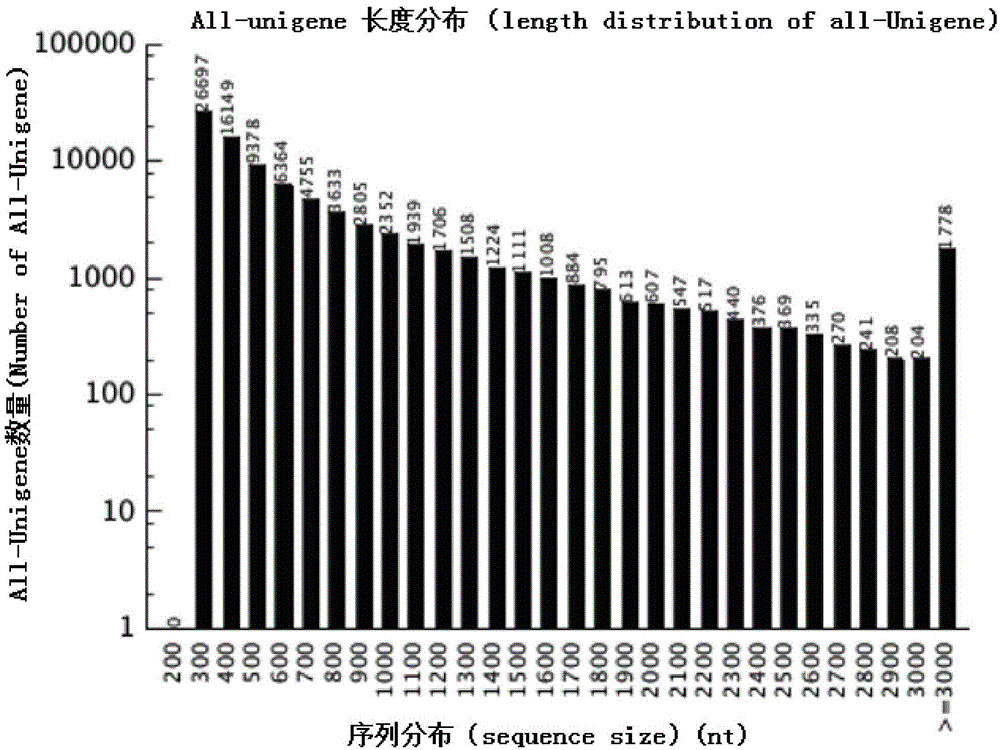
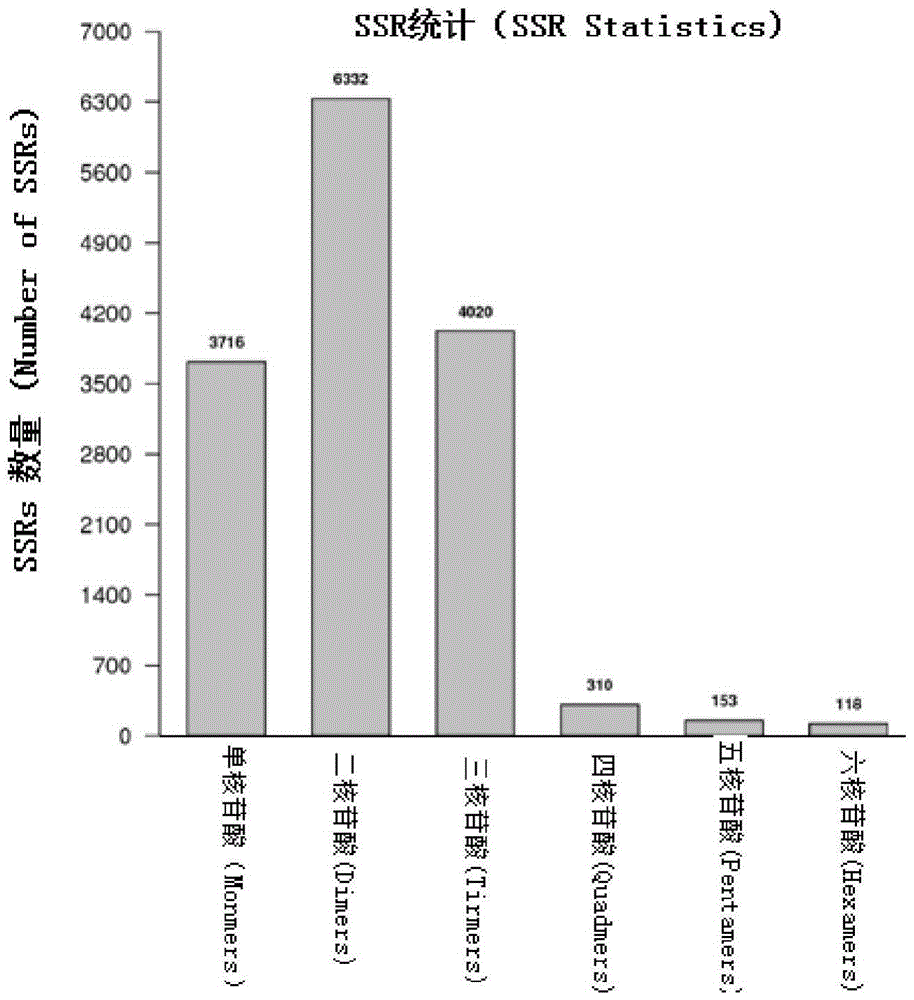
[0025] 2. Acquisition of Unigene and detection of microsatellite loci
[0026] The original ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com