Molecular marker KpnI-1 for indicating and identifying watermelon pulp color and application
A molecular marker, kpni-1 technology, applied in the field of molecular biology, can solve the problems of low polymorphism, few molecular markers, narrow genetic distance of watermelon, etc., and achieve the effect of shortening breeding years and improving breeding efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Primer design and extraction of genomic DNA
[0037] Select the light yellow watermelon line Cream of Saskatchewan (abbreviated as "COS") and the red watermelon line LSW-177 and their crosses to obtain F 1 , F 2 The generation population is the experimental material to verify the CAPS marker to identify the presence of lycopene in watermelon pulp. The result is figure 1 Shown.
[0038] The two watermelon lines COS and LSW-177 were sequenced using high-throughput sequencing technology, and the cds region sequence of the watermelon lycopene β-cyclase gene was obtained through the NCBI website, and the sequence was compared with the sequencing materials. The chromosomal interval in which the cds region sequence of the watermelon lycopene β-cyclase gene exists is analyzed. Within this interval, the base section of the sequencing data with the SNP mutation site is excavated, and the restriction information of the SNP site is analyzed to obtain the existence For CAPS m...
Embodiment 2
[0039] Example 2 Obtaining PCR products
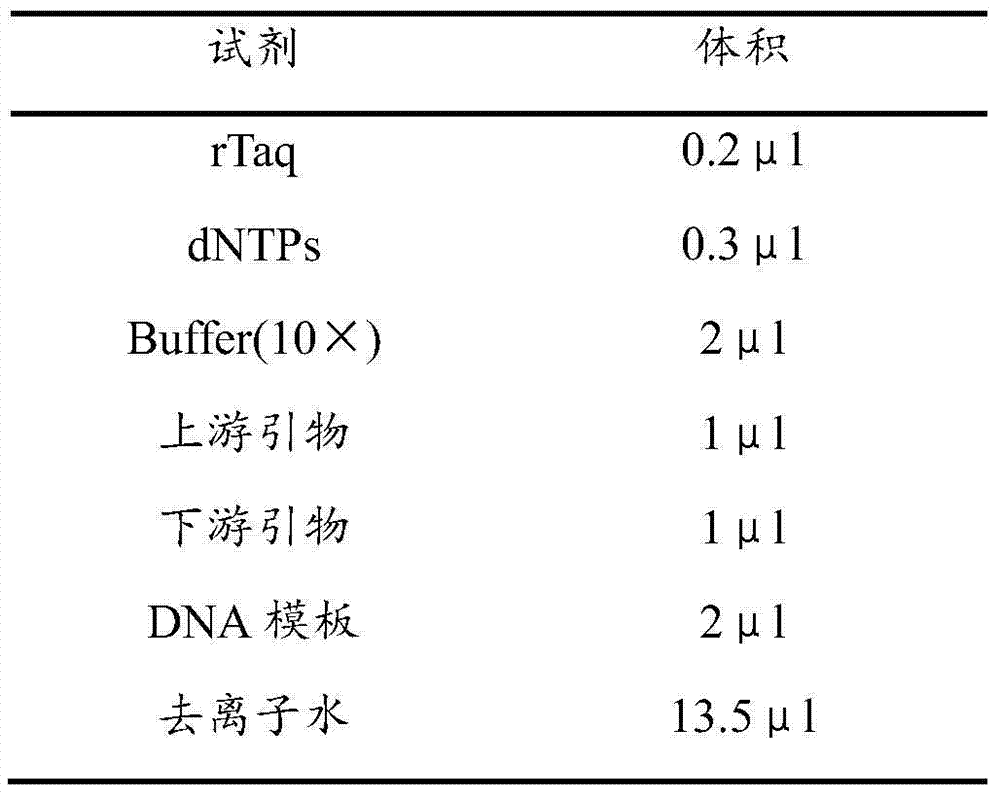
[0040] Using the obtained CAPS primers and genomic DNA to perform PCR reaction, the PCR reaction system is shown in Table 1 (20μl):
[0041] Table 1. PCR reaction system
[0042]
[0043] Using touchdown PCR (TD-PCR) for amplification, the procedure is: 94℃ pre-denaturation for 7 minutes, 94℃ denaturation for 30 seconds, 60℃ annealing for 30 seconds, 0.5℃ per cycle, 72℃ extension for 40s, 30 cycles; 94 Denaturation at ℃ for 30s, annealing at 45℃ for 30s, extension at 72℃ for 40s, 10 cycles; extension at 72℃ for 10min, storage at 4℃.
[0044] Use 1% agarose gel electrophoresis to detect the obtained PCR products, the detection results are as follows figure 1 As shown, LSW-177, COS, F 1 And F 2 Each individual plant amplified a fragment of 517 bp, which was consistent with the expected fragment size.
Embodiment 3
[0045] Example 3 Enzyme digestion verification of PCR products
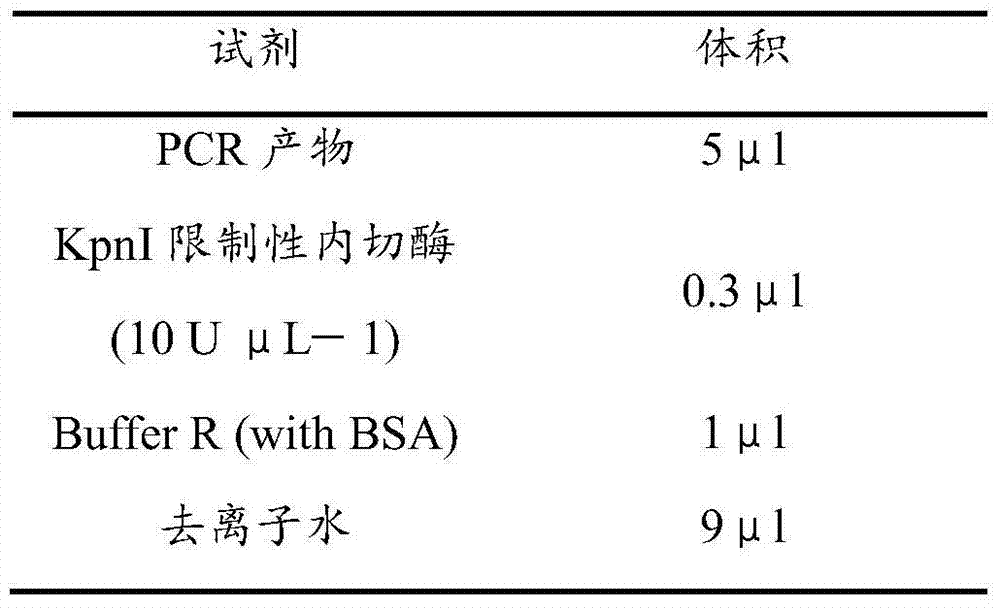
[0046] According to Thermo's restriction endonuclease operation guide, use restriction endonuclease to verify the obtained PCR product. The restriction enzyme digestion system is shown in Table 2: (15.3μl)
[0047] Table 2. Enzyme digestion reaction system
[0048]
[0049] Digestion in a water bath at 37°C overnight, and the digested product is detected by 1% agarose electrophoresis. The detection result is as follows figure 1 Shown. Since LSW-177 has no restriction enzyme site, it is still 517 bp without restriction endonuclease. Due to the restriction enzyme site in COS, it is cut by KpnI to obtain 389 and 128 bp fragments. F 1 The generations showed co-dominant features including not only a 517 bp fragment of LSW-177, but also 389 and 128 bp fragments. In the 18 strains tested F 2 Among the individual plants, 9 plants showed red in the field phenotype, and lycopene could be detected in the pulp tissue. The bands ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com