Quintuple fluorescent PCR detection kit for foodborne pathogenic bacteria
A food-borne pathogenic bacteria and detection kit technology, applied in the biological field, can solve the problems of dye redistribution, limited melting curve, poor stability, etc., and achieve the effect of sensitive automation and low detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Establishment of real-time fluorescent PCR detection method for five kinds of food-borne pathogens
[0038]Extract Staphylococcus aureus, Listeria monocytogenes, Salmonella, Vibrio parahaemolyticus and Shigella standard strain DNA as a template, and use five pairs of specific primers to perform amplification reaction on a real-time fluorescent PCR instrument, 20 μL amplification reaction The system is: template DNA 1 μL, Bio-Rad SsoFast EvaGreen master mix 10 μL, 10 μmol / L nuc upstream and downstream primers 1.4 μL each, 10 μmol / L hlyA upstream and downstream primers 1.2 μL, 10 μmol / L invA upstream and downstream primers 1 μL each, 10 μmol / L 0.8 μL of tlh upstream and downstream primers, 0.8 μL of 10 μmol / L ipaH upstream and downstream primers, and make up to 20 μL with ultrapure water. The amplification reaction conditions are: pre-denaturation at 95°C for 2 minutes, 30 cycles at 95°C for 5s and 62.3°C for 30s, collecting fluorescence during the annealing st...
Embodiment 2
[0039] Embodiment 2: specificity experiment
[0040] Enterobacter sakazakii (ATCC 51329), Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), Yersinia enterocolitica (ATCC 23715), Bacillus cereus (ATCC 11778 ), Vibrio mimicus (ATCC 33653), Vibrio alginolyticus (ATCC 17749) as samples to be tested, Staphylococcus aureus (ATCC 25923), Salmonella typhi (ATCC 50097), Shigella flexneri (ATCC 12022) , Vibrio parahaemolyticus (ATCC 33847) and Listeria monocytogenes (CMCC 54006) were used as positive controls, and ultrapure water was used as negative controls. Enterobacter sakazakii (ATCC 51329), Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), Yersinia enterocolitica (ATCC 23715), Bacillus cereus (ATCC 11778) , Vibrio mimeticum (ATCC 33653), Vibrio alginolyticus (ATCC 17749), Staphylococcus aureus (ATCC 25923), Salmonella typhi (ATCC 50097), Shigella flexneri (ATCC 12022) and Vibrio parahaemolyticus Bacteria (ATCC 33847) was obtained from the Amer...
Embodiment 3
[0041] Embodiment 3: Sensitivity experiment
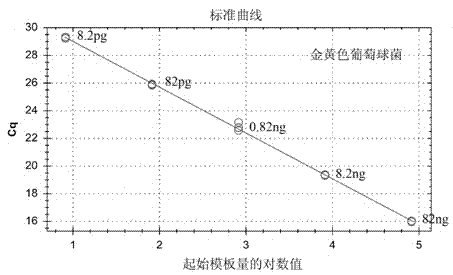
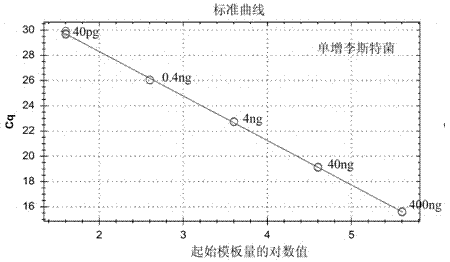
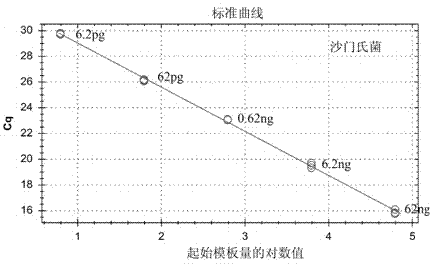
[0042] Using standard strain DNA extracts of Staphylococcus aureus, Listeria monocytogenes, Salmonella, Vibrio parahaemolyticus, and Shigella as templates, dilute with ultrapure water to form a series of dilutions, and take 1 μL template for each dilution The DNA was detected according to the method described above, and three replicate wells were made for each dilution, and the detection was carried out according to the method described in Embodiment 1. The results show that the concentration of each standard strain template DNA has a good linear relationship with the Cq value, and the higher the concentration, the lower the Cq value. For the test results, see Figure 6-10 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com