Method for detecting community structures and abundance of nitrite oxidizing bacteria in wastewater
A technology of nitrite and community structure, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of poor probe specificity, cumbersome steps, and high cost of experimental detection, and achieve strong specificity and wide detection range Broad, comprehensive effect on sequencing sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Primer Design
[0027] The specific amplification forward and reverse primers SEQ ID No.1 and SEQ ID No.2 of the nxrA gene were optimized and designed according to the primers in the Poly et al.2008 literature, and the length of the amplified product was 322bp.
[0028] Select the nxrB gene in the whole genome of Ca. Nitrospira defluvii with the accession number FP929003.1 in the GenBank database, and other partial nxrB gene sequences with a similarity of more than 99% as primer design templates, set the optimal primer parameters, and design specific primers SEQ ID No.3 and SEQ ID No.4, the length of the amplified product of nxrB gene is 506bp.
[0029] nxrA gene forward primer SEQ ID No.1: 5'-GGGTACAAGAAGTGCGTGGA-3';
[0030] nxrA gene reverse primer SEQ ID No.2:
[0031] 5'-CTTGCCGTGGATCTGGGTC-3'
[0032] nxrB gene forward primer SEQ ID No.3: 5'-CAGACCGACGTGTGCGAAAG-3'
[0033] nxrB gene reverse primer SEQ ID No. 4: 5'-TCCACAAGGAACGGAAGGTC-3'.
Embodiment 2
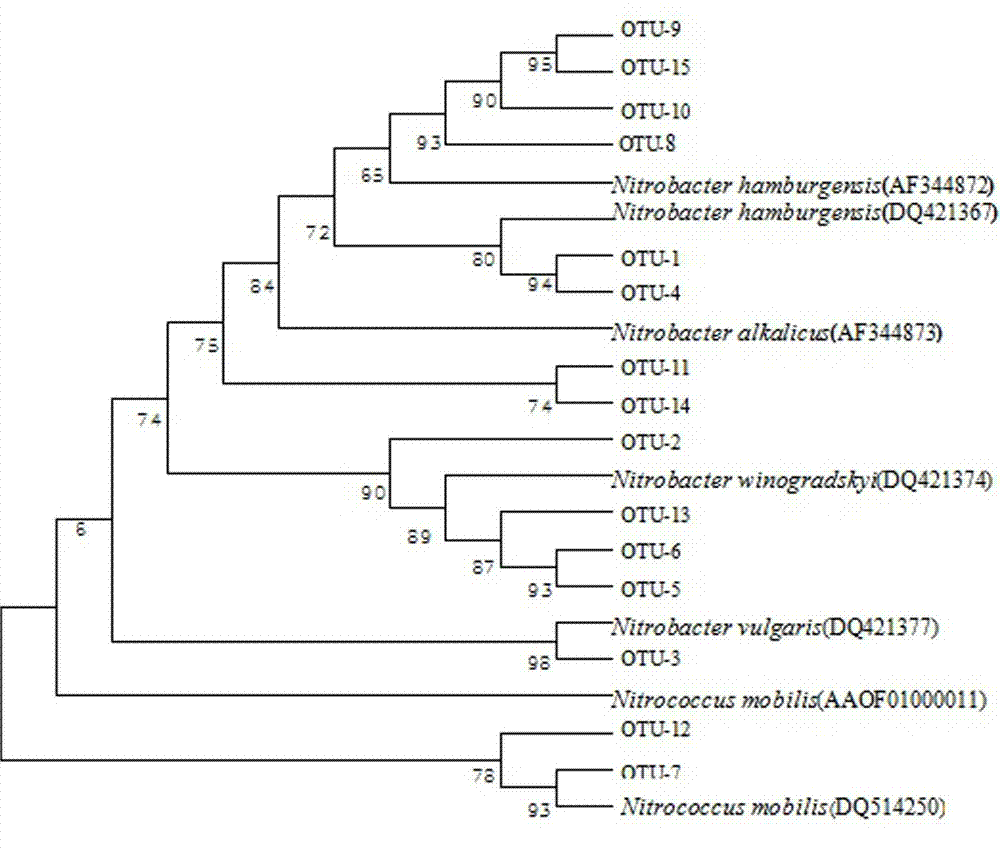
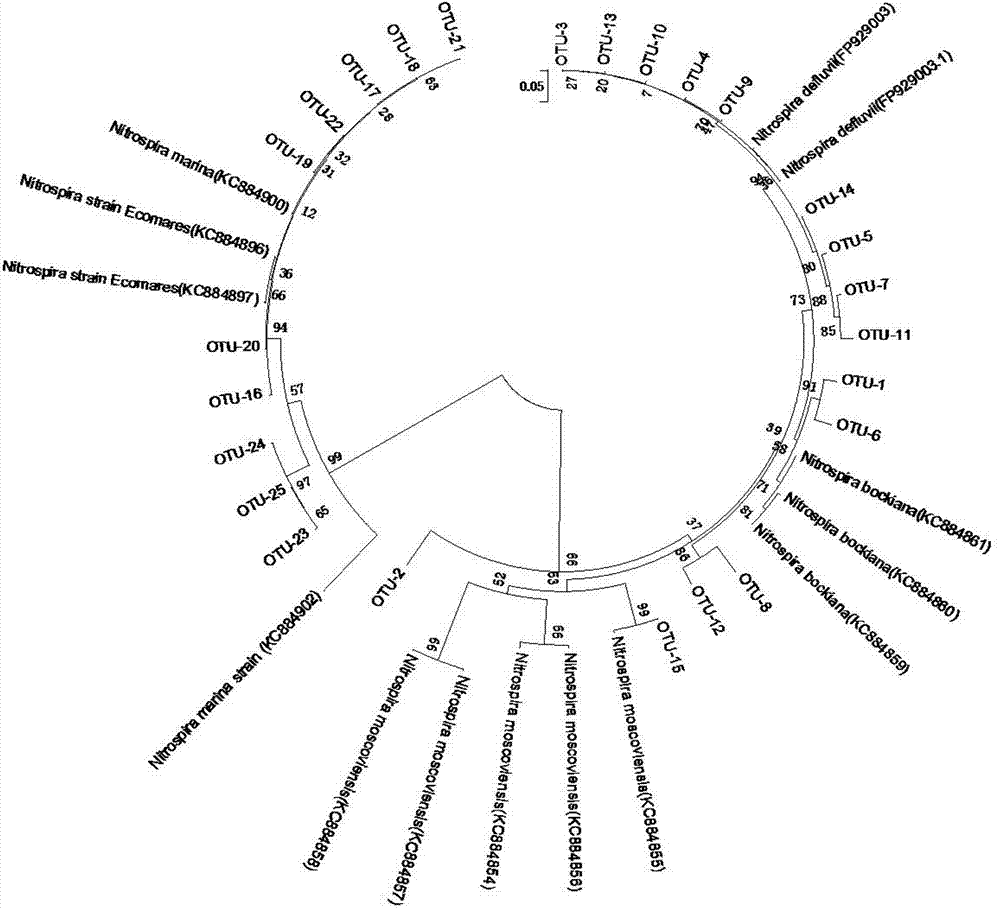
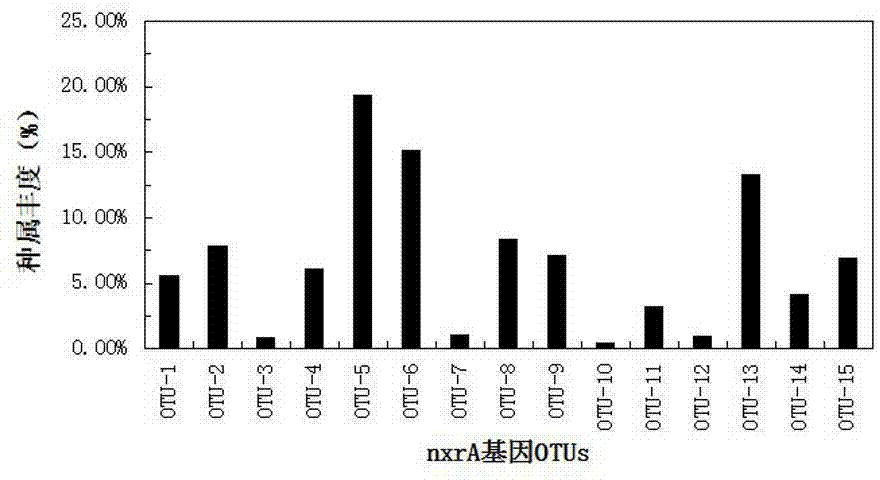
[0034] Example 2 Detection of the community structure and abundance of nitrite oxidizing bacteria in the activated sludge of the aerobic tank of the sewage treatment plant
[0035] (1) Extraction of microbial total DNA in activated sludge
[0036]The sample was taken from the activated sludge in the aerobic tank of a certain urban sewage treatment plant. The FastDNA Soil Spin Extraction Kit produced by MP Biomedicals was used to extract the total DNA from the activated sludge sample. The operation was carried out in strict accordance with the instructions. A luminometer detects the concentration and purity of the total genomic DNA to ensure that the total DNA template is available.
[0037] (2) PCR amplification of nitrite oxidase nxrA and nxrB functional genes
[0038] The PCR reaction systems of nxrA and nxrB genes are both 30 μL, first put the corresponding volume of Taq enzyme, MgCl 2 , forward and reverse primers, deoxyribonucleotide dTNP and sterile double-distilled...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com