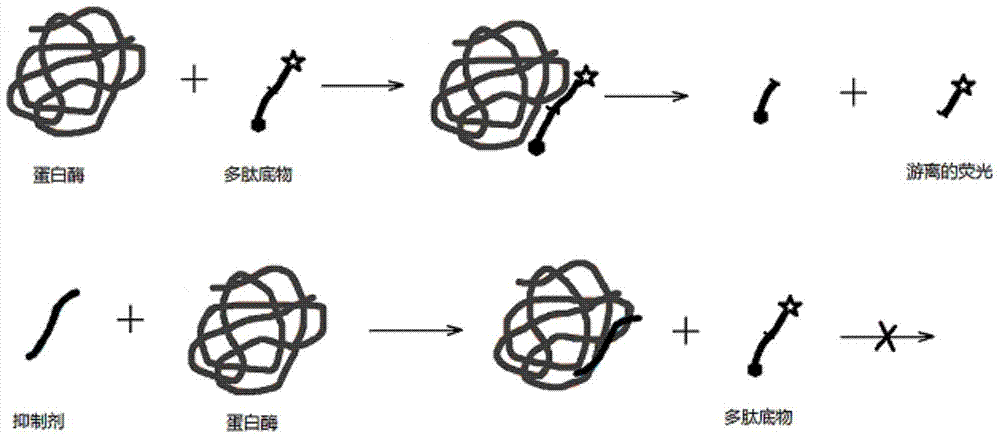
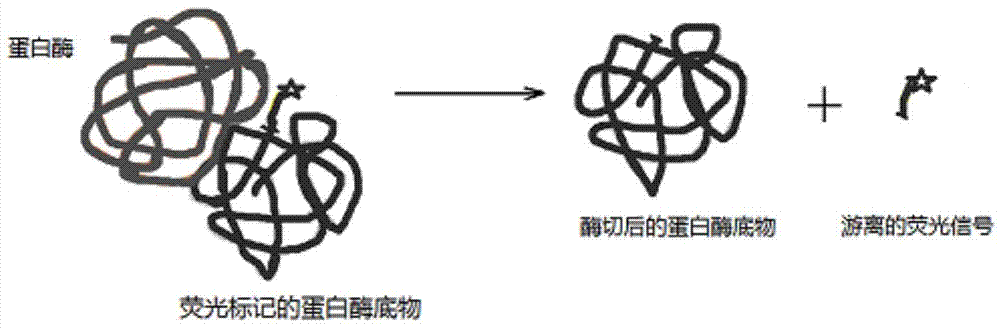
Detection method based on protein substrate and application thereof
A protein substrate and detection method technology, applied in the field of biochemistry, can solve the problems of inability to achieve high throughput, time-consuming, labor-intensive, termination, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0093] Renin is a protease that cleaves Angiotensinogen to produce Angiotensinogen I polypeptide. The Renin-Angiotensinogen system plays an important role in regulating blood pressure. The amino acid sequence of Angiotensinogen was obtained from UniProtKB. To generate an N-terminal fluorescently labeled Angiotensinogen, a cysteine was added to the N-terminal of Agiotensinogen, as in Figure 5 A shown. The amino acid sequence was optimized and synthesized by conventional methods. The gene was cloned into pE-SUMO vector through BsaI and XbaI, and transformed into Escherichia coli BL21 (DE3). This protein is expressed in fusion with SUMO protein, and the fusion protein has a 6×His tag at the N-terminus. The bacteria were cultured. When the OD600 reached 1.0, 0.5 mM IPTG was added to induce expression. The bacteria were resuspended with 25 mM Tris-HCl, 150 mM NaCl, pH 8.0, and disrupted by ultrasonic waves. Subsequently, the expressed protein was purified by Ni affinity chromat...
Embodiment 2
[0097] Beta-secretase (BACE) is one of two proteases that cleaves amyloid precursor protein to produce amyloid polypeptide 1-42. The human APP protein contains 770 amino acids, and the full-length APP contains several domains. The cleavage site of BACE is located between amino acids 671 and 672. The amino acid sequence of APP was obtained from UniProtKB. In this example, the amino acid sequence ranged from 365 to 699 aa. After synthesizing the gene sequence by conventional methods, the gene was cloned into the pTXB1 vector with NdeI and SpeI, and the target protein was directly combined with the contents of the vector. Peptide tag (intein) fusion. The constructed expression vector was transformed into Escherichia coli BL21 (DE3), and the bacteria were cultured. When the OD600 reached 0.6, 0.5 mM IPTG was added to induce expression. The bacteria were resuspended with 25 mM Tris-HCl, 150 mM NaCl, and pH 8.0. Ultrasonic fragmentation was performed, and then the expressed protei...
Embodiment 3
[0100] In this example, 5-TAMRA was labeled at the C-terminus of a shorter form of APP containing one domain (APP550-685) using the same method used to produce APP365-699-(5-TAMRA), as in Figure 12 shown. PCR was used to amplify the gene according to the conventional method, the gene was cloned into pTXB1 vector with NdeI and SpeI, and the target protein was directly fused with the intein tag (intein) carried by the vector. The constructed expression vector was transformed into Escherichia coli BL21 (DE3), and the bacteria were cultured. When the OD600 reached 0.6, 0.5 mM IPTG was added to induce expression. The bacteria were resuspended with 25 mM Tris-HCl, 150 mM NaCl, and pH 8.0. Ultrasonic fragmentation was performed, and then the expressed protein was purified by chitin affinity chromatography. At this time, the purified protein was retained on the chitin column. The resin was rapidly washed with 50mM MESNA in 25mM Tris-HCl, 150mM NaCl, pH 8.0, and then The column was p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com