Probe based on double-signal amplification triggered by target and application of probe
A dual-signal amplification and target technology, applied in the field of dual-signal amplification probes and their detection proteins, can solve the problems of long time, high cost, complicated operation, etc., achieve simple detection methods, reduce background noise, prevent The effect of false positive signal generation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
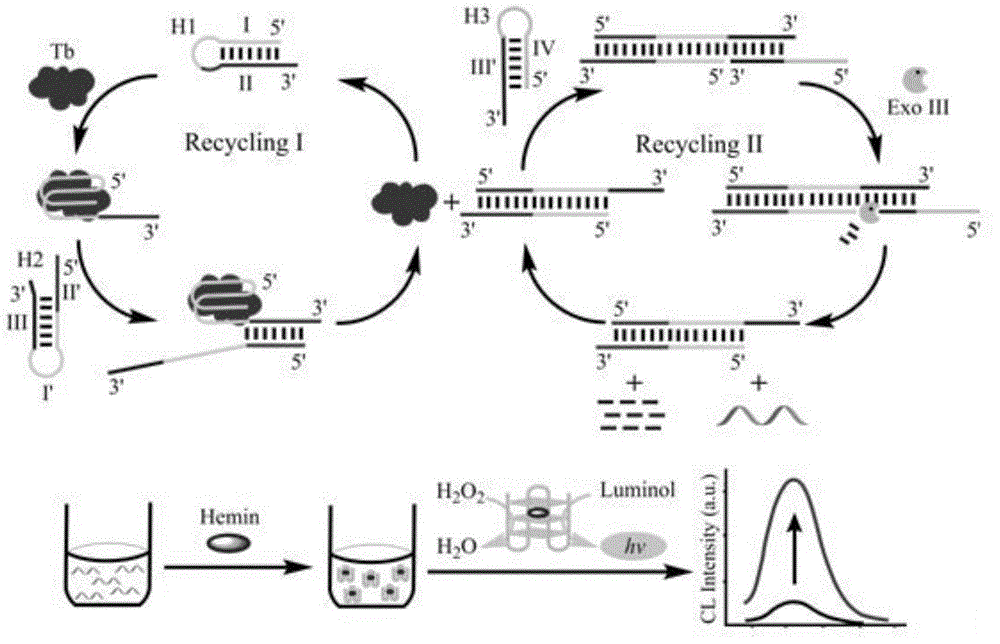
[0048] Example 1 Probe design based on target-triggered dual signal amplification
[0049] The probes for target-triggered dual signal amplification described in this example include: the first hairpin H1, the second hairpin H2 and the third hairpin H3, each of which has a sequence as shown in Table 1 below; The target protein is human thrombin.
[0050] Table 1 Probe sequences based on target-triggered dual signal amplification
[0051]
[0052] The first hairpin, the second hairpin and the third hairpin are all formed by the self-folding of the single-stranded linear molecule, and the complementary base pairing hybridization in the folded region, and the part of the double-stranded structure in the local region is the stem region, the part that does not form a double-strand structure and folds back is a ring region;
[0053] The first hairpin includes: an aptamer region I that can specifically recognize the target protein, and a first stem region II that hybridizes with...
Embodiment 2
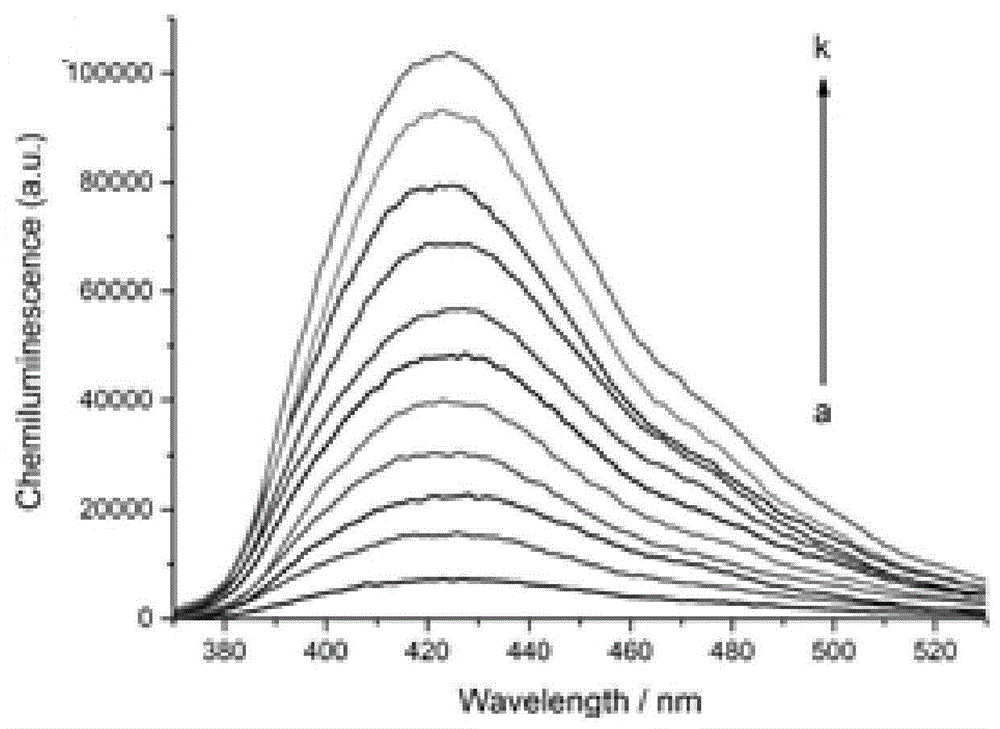
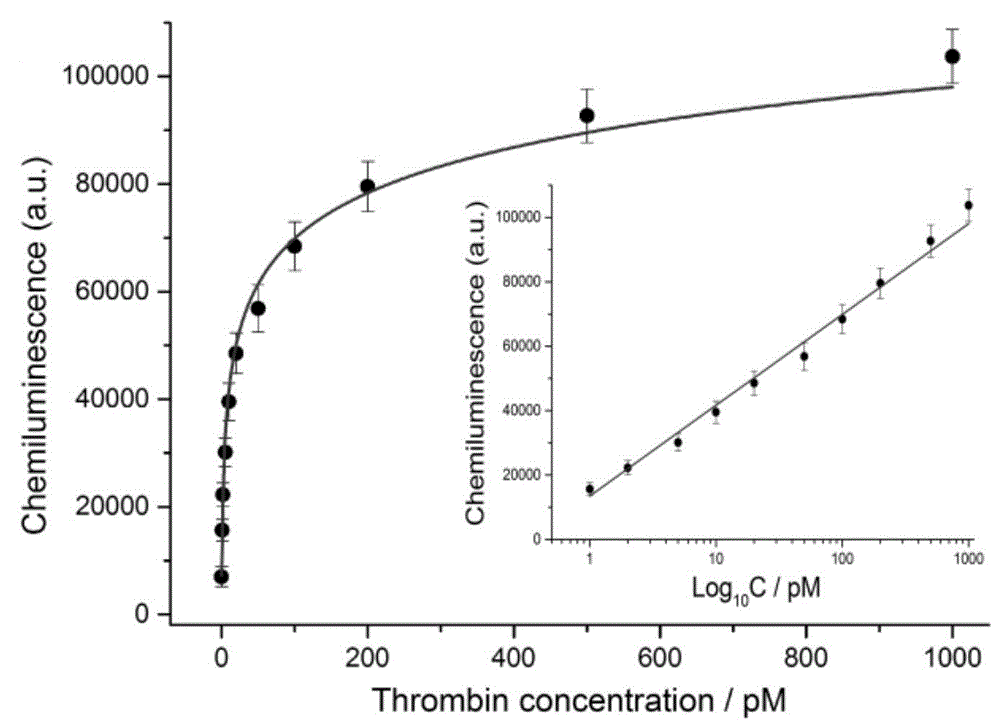
[0068] Example 2 Detection of Target Protein Molecules at Different Concentrations Based on Target-Triggered Dual Signal Amplification Probes
[0069] The method for detecting target protein molecules based on target-triggered dual signal amplification probes of this embodiment includes the following steps:
[0070] First, take the solutions of the first hairpin, the second hairpin, and the third hairpin, heat them to 95° C., keep them for 5 minutes, then slowly lower them to room temperature, and set them aside. Take 10ml of dimethyl sulfoxide (DMSO) and 6.5mg of hemin to prepare 1mM hemin solution, and store it in a dark place at -20°C for future use. 4-Hydroxyethylpiperazineethanesulfonic acid (HEPES) buffer solution was prepared for use, and the buffer solution included: 25 mM HEPES, 200 mM NaCl, 20 mM KCl, 1% DMSO, pH 7.4.
[0071] Prepare Tris-HCl buffer for use, the buffer includes: 20mM Tris-HCl, 200mM NaCl, 20mM KCl, 2mM MgCl 2 , pH value is 7.4. Take human α-throm...
Embodiment 3
[0075] Example 3 Signal Amplification Verification Test of Probes Based on Target-Triggered Double Signal Amplification
[0076] In this example, the following solutions were prepared according to the method in Example 2, and numbered a, b, c, d in sequence:
[0077] Sample a: a solution in which the concentrations of the first hairpin H1, the second hairpin H2, and the third hairpin H3 are all 200 nM;
[0078] Sample b: a solution in which the concentrations of the first hairpin H1, the second hairpin H2, and the third hairpin H3 are all 200 nM, and the concentration of the exonuclease III is 20 U;
[0079] Sample c: a solution in which the concentrations of the first hairpin H1, the second hairpin H2, and the third hairpin H3 are all 200 nM, and the concentration of the target thrombin is 1 nM;
[0080] Sample d: a solution in which the concentrations of the first hairpin H1, the second hairpin H2, and the third hairpin H3 are all 200 nM, the concentration of the exonucleas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com