Fluorescent biosensing method used for detecting low molecular ligand target protein and based on combination of inhibition of click chemistry reaction
A technology of click chemical reaction and small molecule ligand, which is applied in the field of fluorescent biosensing, and achieves wide application prospects, fast and easy operation, and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
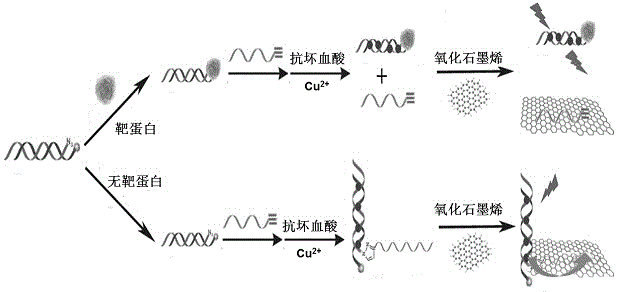
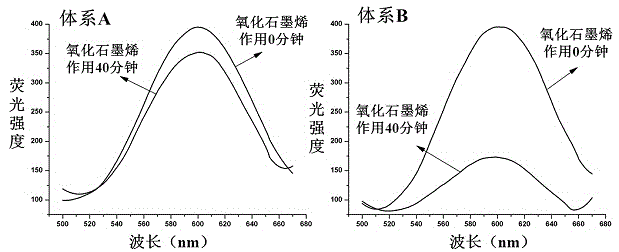
[0027] Embodiment one: The fluorescence characterization of the click chemical reaction, specifically involves the click chemical reaction and the establishment of the "copper nanoparticle-graphene" fluorescence quenching nanoprobe system, the steps are as follows:
[0028]Add 18 μL of 10 μM P1 strand and 18 μL of 10 μM P2 strand to 81 μL of MOPS buffer, mix well and react at room temperature for 2.5 hours to make the P1 and P2 strands complementary hybridize to form a double strand. Subsequently, 90 μL of MOPS buffer, 18 μL of 0 or 10 μM P3 chain, and 5 μL of 100 mM ascorbic acid solution were added to the above solution, and after shaking well, 230 μL of MOPS buffer containing 200 μM copper sulfate was added to the reaction at room temperature. 30 minutes to perform click chemistry and synthesize copper nanoparticles. Finally, 40 μL of 200 μg / mL graphene oxide was added to the above solution, and after the fluorescence was quenched at room temperature for 0 or 40 minutes, ...
Embodiment 2
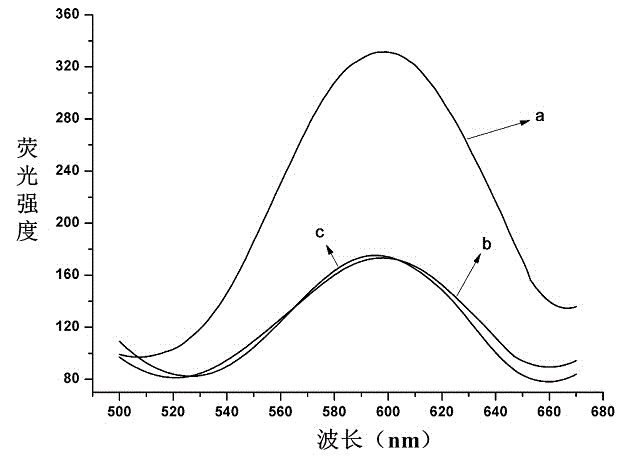
[0035] Embodiment two: Quantitative detection of folate receptors. Folate receptor is a cell surface receptor that binds and transcribes folate molecules (folate) overexpressed in a variety of malignant tumor cells, and its detection is of great significance for the treatment and diagnosis of related cancers. Therefore, the present invention takes the quantitative detection of folate receptor as an example to investigate the feasibility of the established fluorescent biosensing method. It specifically involves the specific recognition between the folic acid molecule modified at the end of the oligonucleotide DNA chain and the folate receptor, the binding inhibition of the click chemical reaction, and the analysis and detection based on the "copper nanoparticle-graphene" fluorescence quenching nanoprobe system, The steps are as follows:
[0036] Take 18 μL of 10 μM P1 chain and 18 μL of 10 μM P2 chain and mix with 81 μL of MOPS buffer. After reacting at room temperature for...
Embodiment 3
[0042] Embodiment three: Detection of folate receptors in complex samples
[0043] In order to study the detection effect of the fluorescent biosensing method established in the present invention in complex samples, we prepared serum samples by dissolving different concentrations of folic acid receptors in human serum (the concentration of folic acid receptors added was 0.8 nM, 3.2 nM and 6.4nM), and perform fluorescence detection according to the steps in Example 2. As shown in Table 1, the recovery rate of folate receptor detection in serum samples was 99.4-106.2%, indicating that this method can meet the needs of complex sample analysis.
[0044] Table 1 The recovery rate of detection of folate receptor in serum samples
[0045]
PUM
| Property | Measurement | Unit |
|---|---|---|
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com