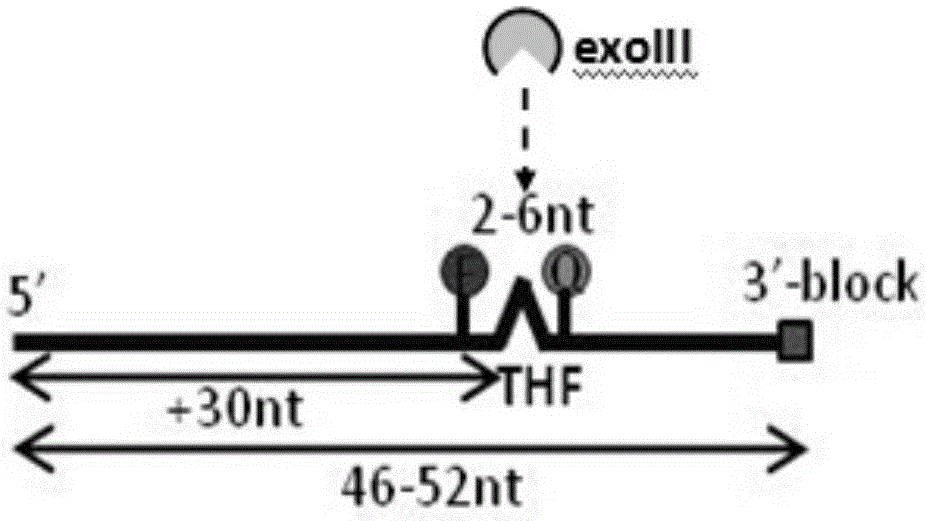
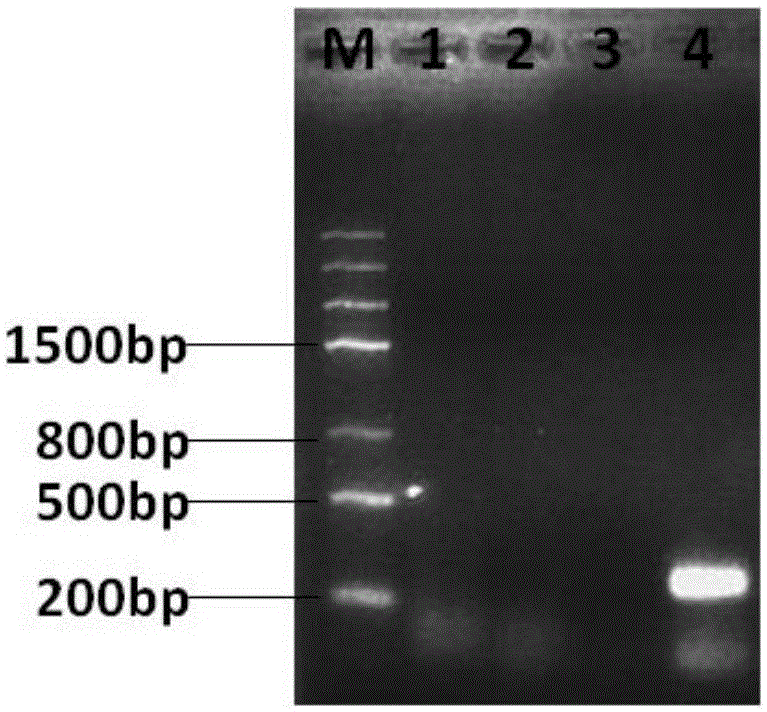
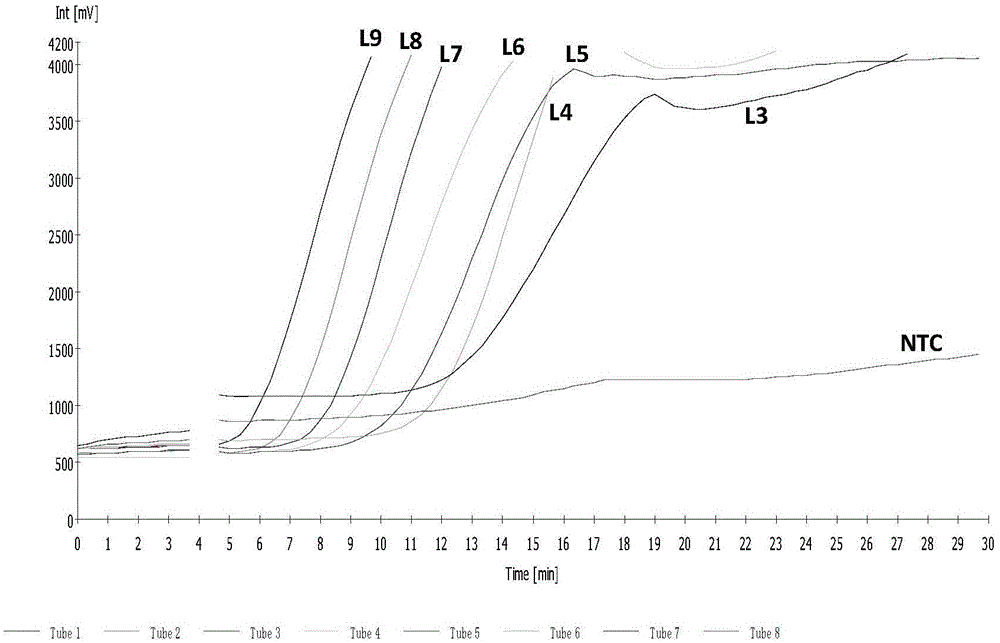
Normal-temperature and constant-temperature nucleic acid amplification method using fluorescence probe
A fluorescent probe, room temperature isothermal technology, applied in the field of molecular biology, can solve the problems of inability to achieve isothermal PCR and imprecise product quantification, and achieve the effect of increasing the detection accuracy and the detection rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] This example is used to illustrate the rapid fluorescence reaction at room temperature and constant temperature performed on the Twista instrument and the multifunctional fluorescent microplate reader BMG Omeg.
[0048] 1. According to NCBI GenBank and related literature reports, primers were designed based on the soybean internal reference gene EF 1b, and a pair of optimal primer pairs (SEQ ID No.6 / 7) was screened out from many primer pairs through experimental optimization. The increased fragment is 235bp (SEQ ID No.8) ( figure 2 ). Subsequently, the fragment was cloned into the plasmid pMD18-T to obtain the desired recombinant plasmid pMD18T-EF1b, which was used as a template for fluorescence detection.
[0049] EF1b-F: 5'-TCACACACACACATATAGAAAGAGAGAGAC-3' (SEQ ID No.6)
[0050] EF1b-R: 5'-CTGCTCCAGTTAGTTCATATAAGAGATAGG-3' (SEQ ID No. 7)
[0051] SEQ ID No. 8:
[0052] 5’-TCACACACACACATATAGAAAGAGAGAGACAATGGTTGTCAGCATCGTCCATTTCTTTTTAGACCAAAGATTTTAATGCAGTCAACTCTGC...
Embodiment 2
[0064] This example is used to illustrate the fluorescence detection of the MTB insertion sequence IS6110 gene on the Twista instrument.
[0065] 1. According to NCBI GenBank and related literature reports, a 523bp sequence (SEQ ID No.10) was selected from the insertion sequence IS6110 of MTB for gene synthesis, and cloned into the vector pUC57 to prepare a recombinant plasmid pUC57-6110. Template for fluorescence detection.
[0066] SEQ ID No. 10:
[0067] 5’-CTCCGACCGACGGTTGGATGCCTGCCTCGGCGAGCCGCTCGCTGAACCGGATCGATGTGTACTGAGATCCCCTATCCGTATGGTGGATAACGTCTTTCAGGTCGAGTACGCCTTCTTGTTGGCGGGTCCAGATGGCTTGCTCGATCGCGTCGAGGACCATGGAGGTGGCCATCGTGGAAGCGACCCGCCAGCCCAGGATCCTGCGAGCGTAGGCGTCGGTGACAAAGGCCACGTAGGCGAACCCTGCCCAGGTCGACACATAGGTGAGGTCTGCTACCCACAGCCGGTTAGGTGCTGGTGGTCCGAAGCGGCGCTGGACGAGATCGGCGGGACGGGCTGTGGCCGGATCAGCGATCGTGGTCCTGCGGGCTTTGCCGCGGGTGGTCCCGGACAGGCCGAGTTTGGTCATCAGCCGTTCGACGGTGCATCTGGCCACCTCGATGCCCTCACGGTTCAGGGTTAGCCACACTTTGCGGGCACCGTAAACACCGTAGTTGGCGGCGTGGACGCGGCTGATGTGCTC-3’ ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com