A molecular marker method and its primer pair that can predict and identify the curl degree of sheep wool
A technology of curling and sheep, applied in the field of animal molecular genetics, achieves the effect of high precision, simple operation and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Obtaining Genomic DNA of Chinese Merino Sheep
[0033] The sheep ear tissues were collected and stored at -20°C for future use. The sheep genomic DNA was extracted by conventional phenol / chloroform method. The specific method is as follows:
[0034] (1) Take 5 g of ear tissue from Chinese Merino sheep (Xinjiang Army Reclamation type), remove the connective tissue, clean and disinfect the tissue block with 70% alcohol, put it in an Eppendorf tube, cut it with scissors, or grind it to pieces;
[0035] (2) After the alcohol in the Eppendorf tube is completely evaporated, add 700 μL of separation buffer, suspend the chopped tissue, add 5.0 μL of proteinase K (20 mg / mL), and act at 55 ° C for 8-12 hours until there are no tissue pieces;
[0036] (3) Take out the digested tissue fluid, add an equal amount of saturated phenol in the tissue fluid, mix for 10 minutes, centrifuge at 12,000 r / m at 4°C for 10 minutes;
[0037] (4) Take the supernatant, add the same amount of phe...
Embodiment 2
[0043] Obtaining of Enzymatic Digestion Product of Sheep SHCBP1 Gene
[0044] According to the SHCBP1 gene G14408374T site in the sheep genome, a pair of primers SHCBP1-P1-F and SHCBP1-P1-R were designed, and then the sheep genome DNA was amplified by PCR to obtain PCR amplification products, and then the endonuclease Taq I enzyme Cut the PCR amplified product to obtain the digested product.
[0045] The primer sequences are as follows:
[0046] SHCBP1-P1-F: 5'-TGATTCAATGCGCAACTGGTA-3'
[0047]SHCBP1-P1-R: 5'-AGACATGAAGCAAAGTATGATAGCAG-3'.
[0048] The PCR amplification system is as follows:
[0049]
[0050] PCR amplification conditions were: 94°C pre-denaturation for 5 min; 94°C denaturation for 30 s, 53.8°C annealing for 25 s, 72°C extension for 30 s, a total of 40 cycles; 72°C final extension for 10 min.
[0051] The enzyme digestion reaction system is as follows:
[0052] 10×Buffer Taq I 1μL
[0053] Endonuclease Taq I 1 μL
[0054] PCR product 10μL
[0055] Th...
Embodiment 3
[0057] Determination of Sheep SHCBP1 Genotype
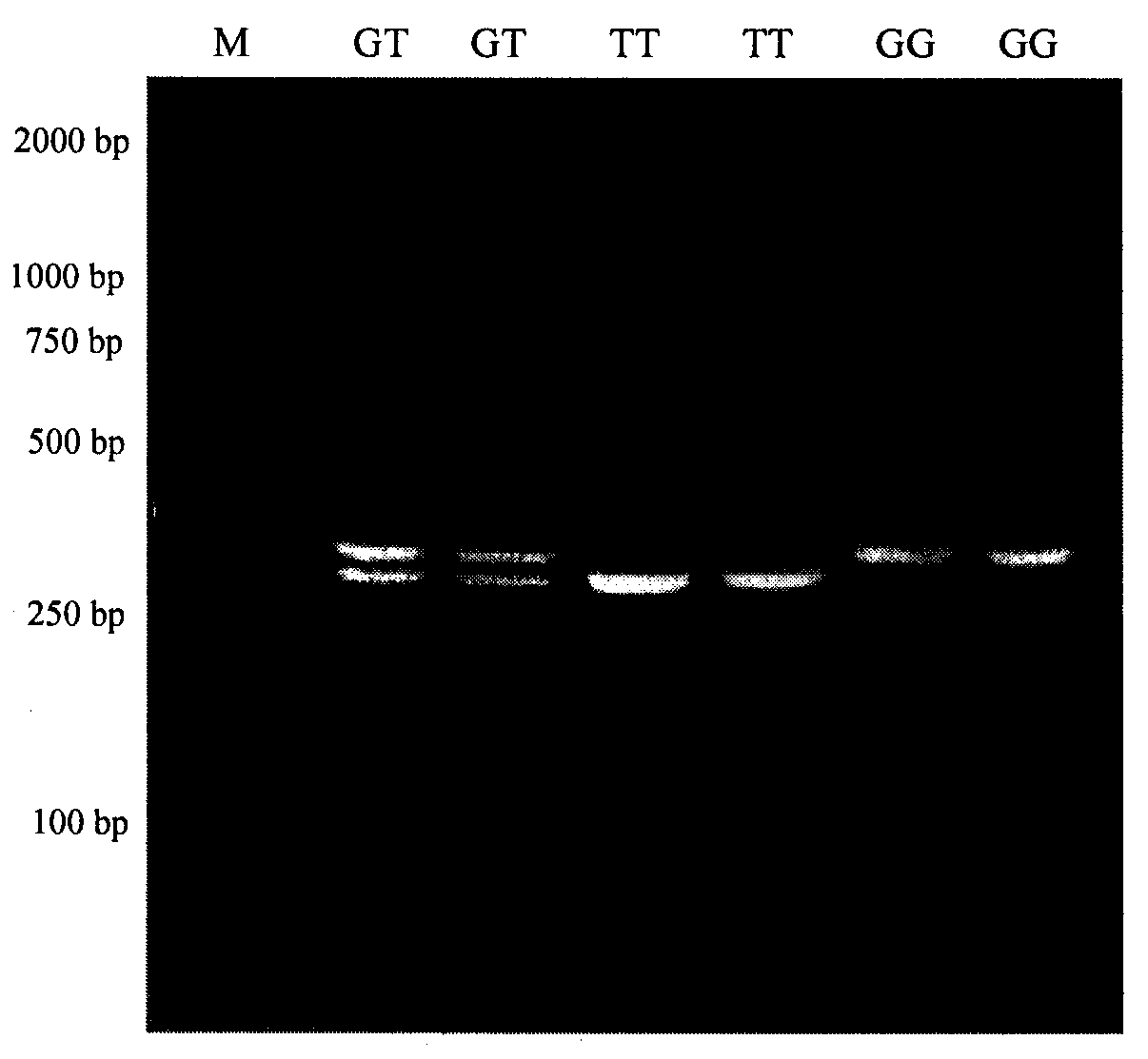
[0058] Use agarose gel with a concentration of 2% to 3% to carry out electrophoresis separation of the digested products in Example 1, and determine the genotype according to the results of the electrophoresis separation. The criteria for determination: (1) electrophoresis presents two bands, the size of which is 309bp and 36bp, when the G14408374T site mutation in the 14th exon region of the sheep SHCBP1 gene is mutated, the PCR amplification product can be completely cut by Taq I enzyme, and it is named as the TT genotype; (2) Electrophoresis shows a band, the size If it is 345bp, when the G14408374T site in the 14th exon region of the sheep SHCBP1 gene is not mutated, the PCR amplification product cannot be digested by Taq I, and it is named GG genotype; (3) Electrophoresis shows three bands, the size is 345bp , 309bp and 36bp, the G14408374T site in the 14th exon region of the sheep SHCBP1 gene is in a heterozygous state, and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com