DNA (deoxyribonucleic acid) phosphorothioation modifier gene cluster
A technology for phosphorylation and modification of genes, which is applied in the field of DNA phosphorylation modification of gene clusters, and can solve problems such as no discovery.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1: Preliminary inference of CpsC phosphorylation gene cluster
[0055] Download the genome file of marine halophilic Vibrio FF75 from the NCBI website, and upload it to the Rapidannotationusingsubsystemtechnology website for genome annotation. After the analysis is complete, use thioredoxin (thioredoxin, PAPS reductase) and cysteinedesulfurase (cysteine desulfurase) as keywords to search for all about 4,500 genes. The obtained candidate genes were compared upstream and downstream one by one, and a gene cluster composed of three genes with a length of about 6 kb was found, and the third gene was PAPS reductase. At the same time, it was found that there is a gene encoding cysteine desulfurase at about 7 kb upstream of the gene cluster. Other genes in and around this region do not belong to any sulfur metabolism system, so it is inferred that this region is very likely to be a CpsC phosphorylation-modified gene cluster encoding DNA. These 4 genes are named as v...
Embodiment 2
[0056] Example 2: Proof of gene knockout vpt1, vpt2, vpt3 and vpt4 Vibrio halophilus Vibriocyclitrophicus Essential genes for phosphorylation of FF75DNA
[0057] According to the genome sequence of marine halophilus Vibrio FF75, the primers corresponding to the four genes were designed. Using the extracted total DNA of Vibrio FF75 as a template, PCR on the left and right arms of each gene, the second PCR synthesis of left and right arm conjoined, the conjoined are about 1.5kb. The left and right arms of the TA clone were joined together for sequencing. After the result is correct, use the SpeI and SacI double enzyme digestion system to process the recombinant T vector to obtain the left and right arm conjoined fragments; use the same endonuclease combination to process the suicide vector pJC4 plasmid to obtain a double-nicked linear plasmid band. Those are connected by T4 ligase.
[0058] Convert the connected product into E.coli WM6024 (the bacterium is DAP auxotrophic), spr...
Embodiment 3
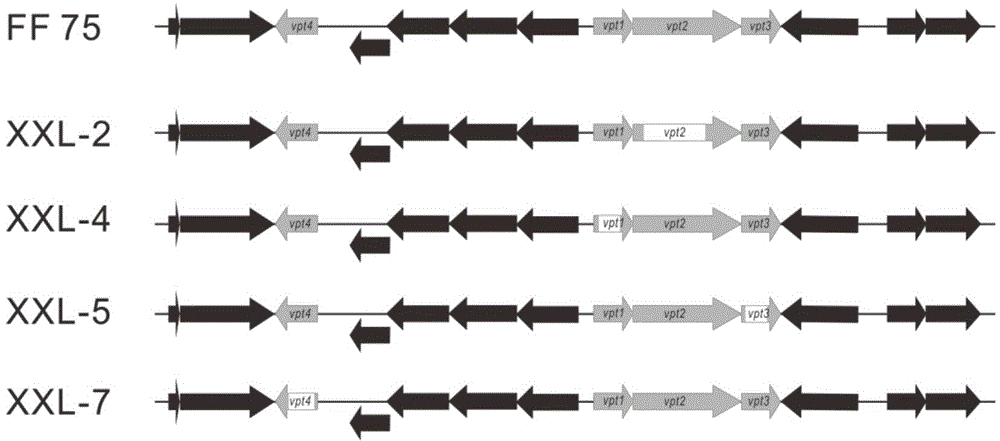
[0061] Example 3: Marine halophilic Vibrio Vibriocyclitrophicus Boundary determination of FF75 DNA phosphorylation modified gene cluster
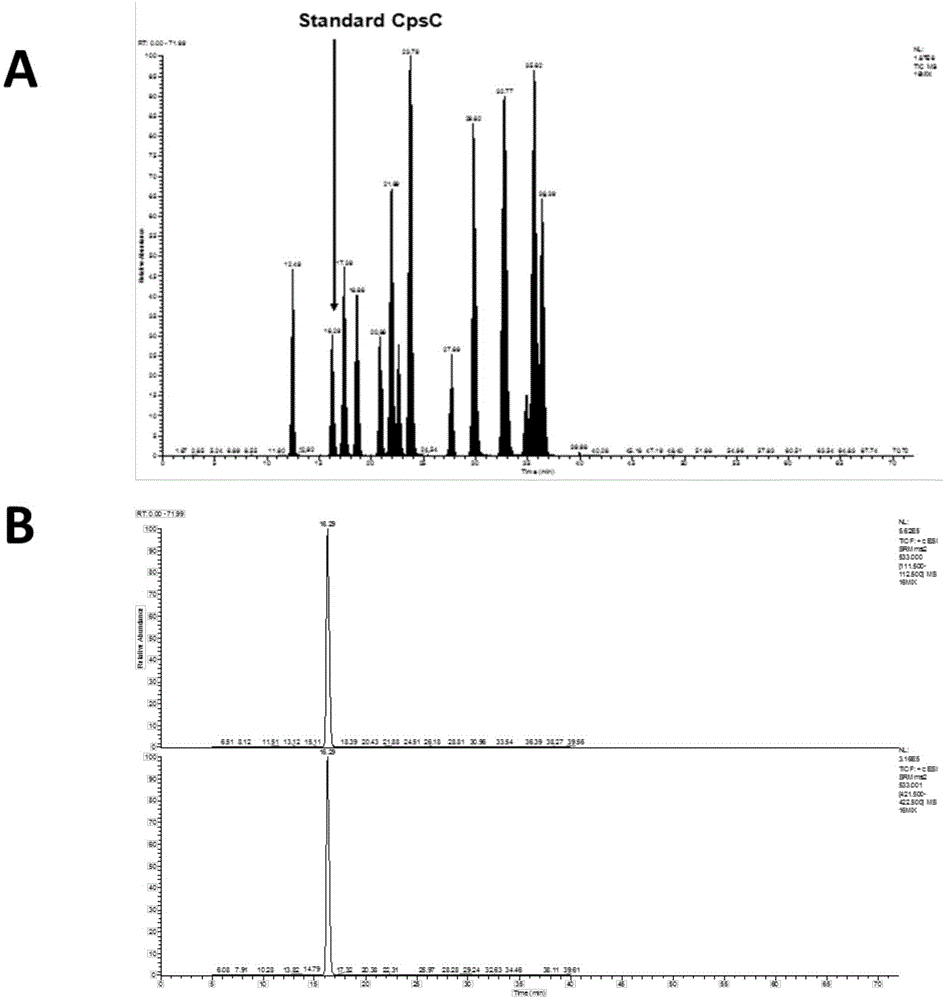
[0062] The mutant strains XXL-4, XXL-2, XXL-5, XXL-7 in Example 2 (knockout respectively vpt1 , vpt2 , vpt3 , vpt4 ) Loss of DNA phosphorylation modification, so according to the gene knockout method in Example 2, the upstream and downstream were knocked out to obtain mutant XXL-1 (knockout 1585) and mutant XXL-3 (knockout Except for 1587-1590), XXL-6 (knockout 1594), tested by LC-MS / MS, experiments show that mutants XXL-1, XXL-3 and XXL-6 still have DNA phosphorylation modification (see respectively under Figure 5 , 6 , 7), therefore, Vibrio Vibriocyclitrophicus The upstream boundary of the FF75 DNA phosphorylation modification gene cluster was determined between 1585 and vpt4 Between, the downstream boundary is determined at vpt3 And 1594, vpt4 with vpt1 There are 1587-1590 between. When knocked out in Example 2 vpt1 , vpt2 , ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com