P genome specific Gypsy retrotransposon and application thereof
A kind of specific, specific primer pair technology, applied in the direction of recombinant DNA technology, microbial assay/inspection, DNA/RNA fragment, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
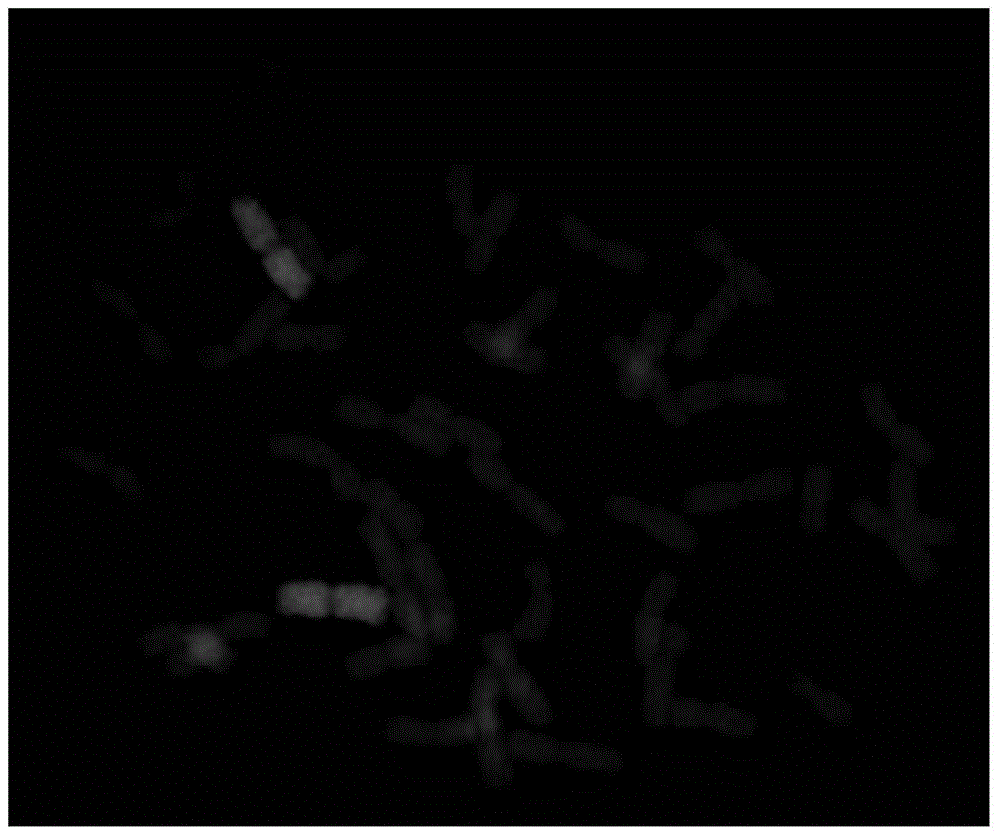
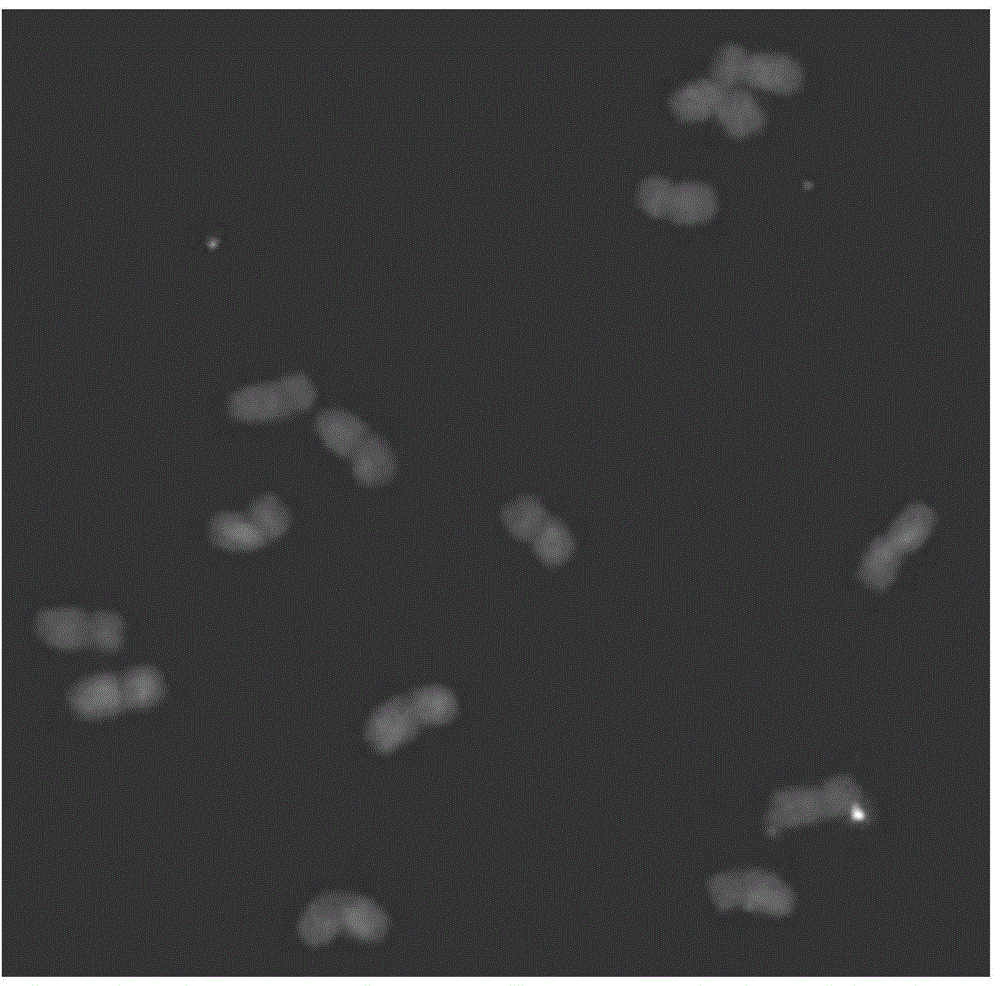
[0065] Example 1, the discovery of pAcPR1 and its distribution characteristics on the chromosome of wheatgrass
[0066] 1. Discovery of pAcPR1
[0067] The 6P short arm DNA sequence fragment was obtained by microdissection and DOP-PCR using wheat-Agropyron 6P short arm system as material. These sequences were connected to the pMD19-T vector and introduced into Escherichia coli to obtain a DNA sequence fragment library of the 6P short arm. Differential DNA clones between wheatgrass Z559 and wheat Fukuho were screened by dot hybridization and sequenced.
[0068] According to BLAST analysis, there is a DNA fragment with a length of 1793bp and a GC content of 37.9%. The DNA fragment is named pAcPR1, as shown in sequence 1 in the sequence listing.
[0069] pAcPR1 has the most sequence similarity (73%) to a long terminal (LTR) Gypsy-like retrotransposon in the wheat 3B-specific BAC library, which is part of cgt0464b.
[0070] pAcPR1 is a new sequence.
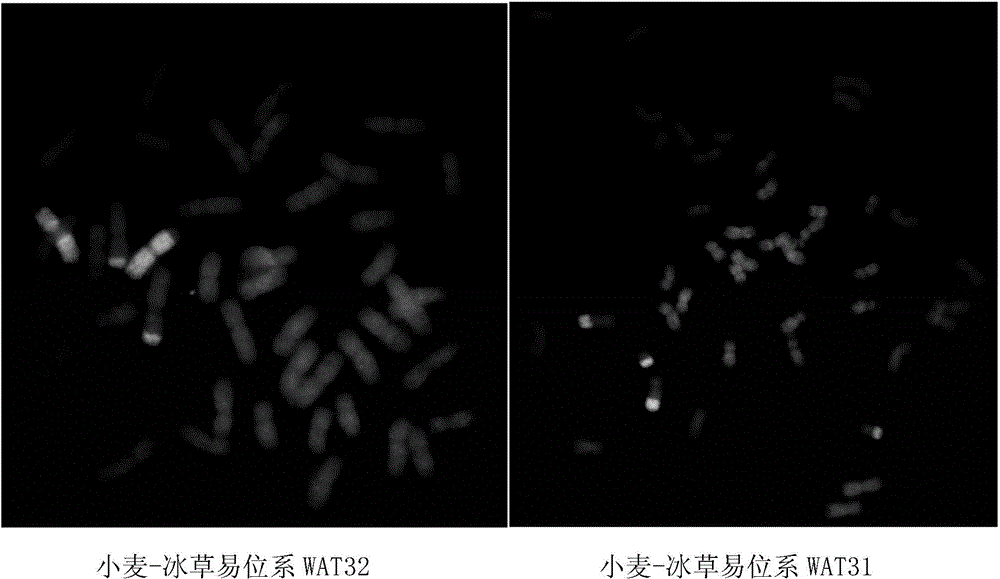
[0071] 2. The distributio...
Embodiment 2
[0077] Embodiment 2, development and application of specific primer pair
[0078] 1. Based on the pAcPR1 sequence, a specific primer pair was designed as follows:
[0079] AcPR1F (upstream primer, sequence 2 in the sequence listing): 5'-CCCCTTCATTAAGGTATTGTTCC-3';
[0080] AcPR1R (downstream primer, sequence 3 in the sequence listing): 5'-CTTGTCCACATGTTGTGTGCTAT-3'.
[0081] 2. Extract the genomic DNA of each sample to be tested respectively.
[0082] 3. Using each genomic DNA as a template, perform PCR amplification using the specific primer pair obtained in step 1 to obtain PCR amplification products.
[0083] 4. Carry out 1% agarose gel electrophoresis for each PCR amplification product obtained in step 3, the results are shown in Figure 4 .
[0084] Figure 4 Among them, M corresponds to marker, 1 corresponds to Wheatgrass Z559, 2 corresponds to wheat Fukuho, 3 corresponds to wheat-Wheatgrass 6P disomy addition line 4844-12, 4 corresponds to wheat-Wheatgrass addition l...
Embodiment 3
[0087] Embodiment 3, the specificity of specific primer pair
[0088] 1. Extract the genomic DNA of each sample to be tested.
[0089] 2. Using each genomic DNA as a template, PCR amplification is carried out with specific primer pairs composed of AcPR1F and AcPR1R to obtain PCR amplification products.
[0090] 3. Perform 1% agarose gel electrophoresis on each PCR amplification product obtained in step 2, the results are shown in Figure 5 .
[0091] Figure 5 Among them, M corresponds to marker, 1 corresponds to wheatgrass Z1842, 2 corresponds to wheatgrass Z559, 3 corresponds to wheatgrass Z1750, 4 corresponds to wheat Chinese spring, 5 corresponds to wheat Fukuho, and 6 corresponds to cultivating einkorn wheat MO 4 , 7 corresponds to durum wheat Jing DR 3 , 8 corresponds to Timofeywheat TI 1 , 9 corresponds to Ae14, 10 corresponds to Y93, 11 corresponds to Y258, 12 corresponds to Ae49, 13 corresponds to Y39, 14 corresponds to RM2161, 15 Corresponds to the cultivated b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com