PCR-HRM primer and method for quickly distinguishing DHAV-1 from DHAV-3
A PCR-HRM and duck hepatitis A virus technology, which is applied in the field of primers for quickly distinguishing duck hepatitis A virus type 1 and type 3 PCR-HRM, can solve the problems of difficulty in differential diagnosis, sensitivity of test results, and restrictions on popularization and application. , to achieve the effect of shortening detection time, good specificity and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Embodiment 1 distinguishes the screening of duck hepatitis A virus type 1 and type 3 PCR-HRM primers
[0059] PCR-HRM primers:
[0060] After screening a large number of designed primers, it was found that the base sequences of the primers SEQIDNO: 1 and SEQ ID NO: 2 have the best effect on the PCR-HRM method for distinguishing DHAV-1 and DHAV-3, and the base sequences are as follows.
[0061] DHAV-P1: 5'-TAGTGTTGTGGGATACCC-3' (SEQ ID NO: 1);
[0062] DHAV-P2: 5'-GTGGGTGTTTTACGTGTACTC-3' (SEQ ID NO: 2).
Embodiment 2
[0063] Embodiment 2 Establishment of a kind of PCR-HRM method for distinguishing duck hepatitis A virus type 1 and type 3
[0064] (1) Extraction and reverse transcription of DHAV-1 and DHAV-3 RNA
[0065] Use the kit TakaraMiniBESTViralRNA / DNAExtractionKitVer.4.0 to extract the RNA in the disease sample, and use TakaraReverseTranscriptaseM-MLV to reverse transcribe the RNA into cDNA.
[0066] (2) Preparation of positive plasmid samples
[0067] The purified DNA of DHAV-1 and DHAV-3 were respectively connected to the pMD-18T vector with a kit from Takara Company, and the positive clones obtained by screening through ampicillin resistance screening, colony PCR and sequencing were positive plasmid samples.
[0068] (3) PCR-HRM operation steps for positive plasmid samples
[0069] Using the three positive plasmid samples obtained above as DNA templates, the PCR-HRM amplification reaction and analysis were carried out respectively;
[0070] Vazyme2×TaqPlusMasterMix5μl
[007...
Embodiment 3
[0081] Embodiment 3 distinguishes the specificity test of duck hepatitis A virus type 1 and type 3 PCR-HRM method
[0082] (1) Extraction of viral RNA or DNA from the sample and reverse transcription: the method is the same as in Example 1 (1) Viral nucleic acid extraction method and RNA reverse transcription.
[0083] (2) Use the established PCR-HRM method to amplify the cDNA of DHAV-1 and DHAV-3 by PCR, and use the nucleic acids of GPV, MDPV, MDRV, NDV, DEV, and DTMUV as control samples to test the specificity of the method . Amplification reaction of PCR-HRM: the method is the same as the reaction system and reaction procedure in Example 1 (2).
[0084] (3) Analysis of specificity test PCR-HRM results
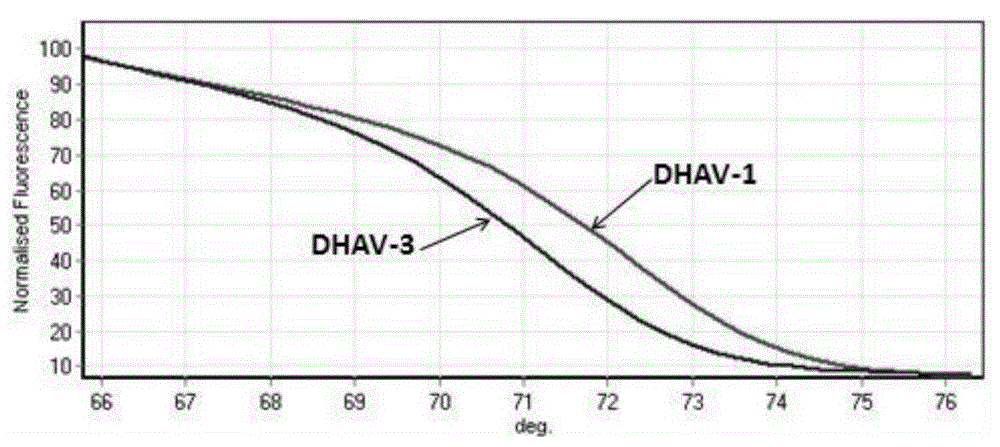
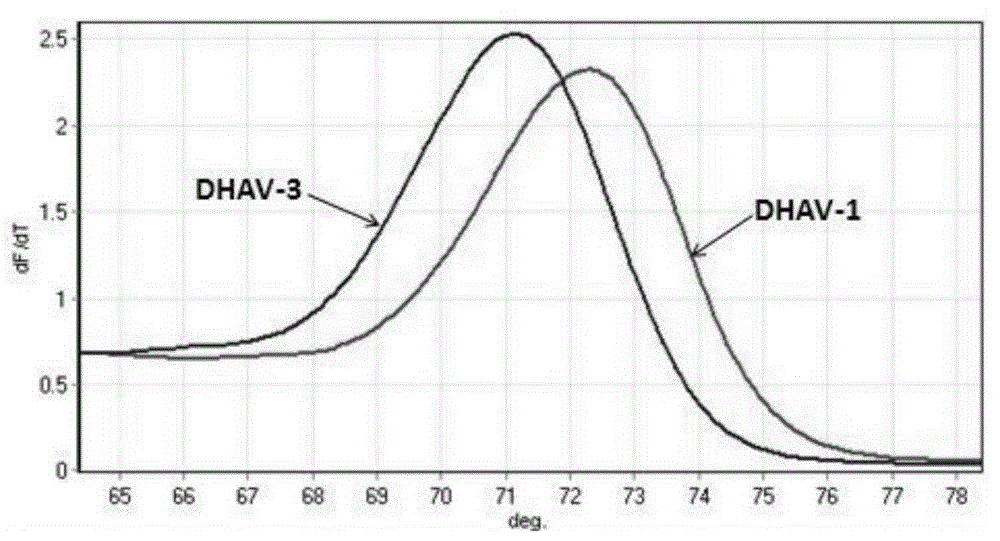
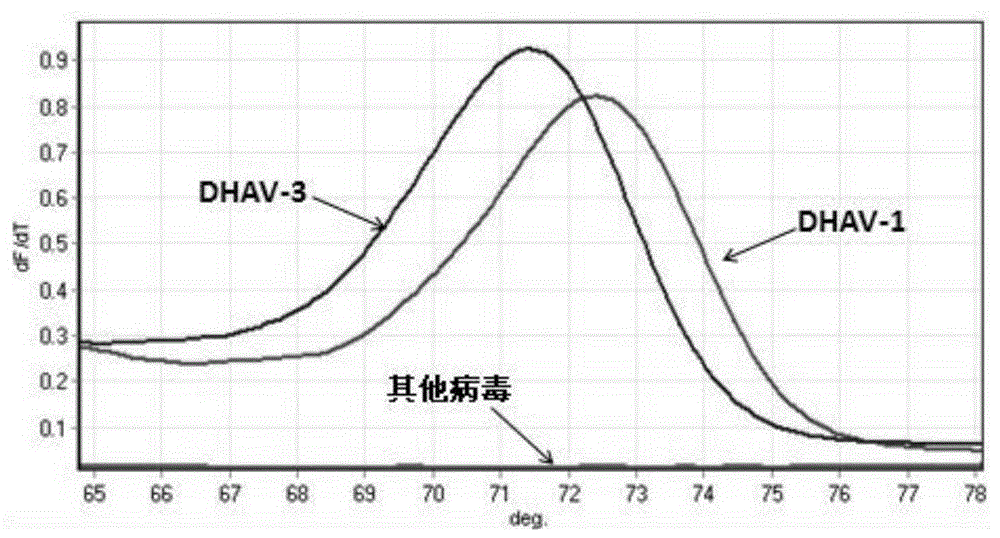
[0085] The PCR amplification product was analyzed with Rotor-GeneQ analyzer, and the results were as follows: image 3 shown.
[0086] From image 3 It can be seen from the peak-shaped melting curve graph shown that the DHAV-1 and DHAV-3 positive samples have the same...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com