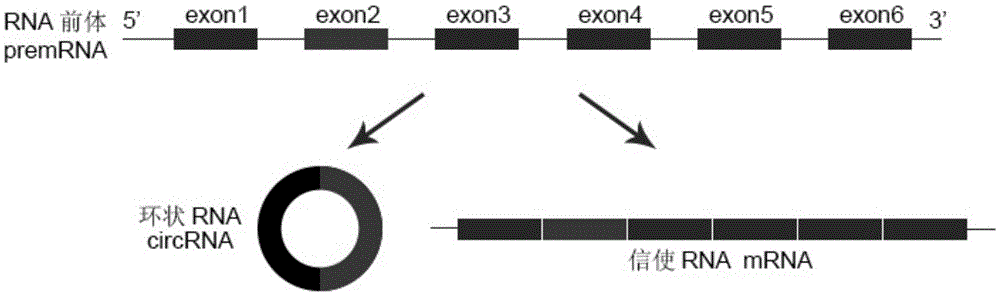
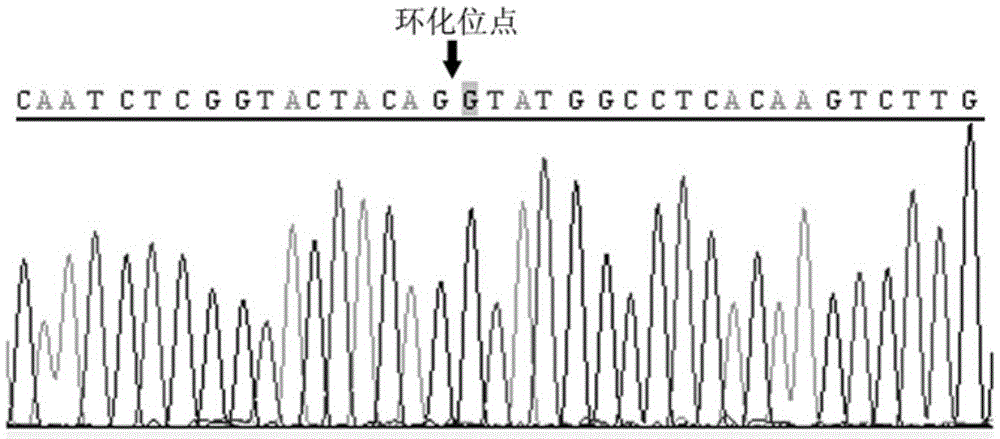
Cyclic RNA circ-ZKSCAN1 use
A circular and versatile technology, applied in the field of circular RNA, can solve the problems of lack of research on liver cancer, achieve the effect of increased cell migration rate and cell invasion ability, and accurate diagnosis of liver cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
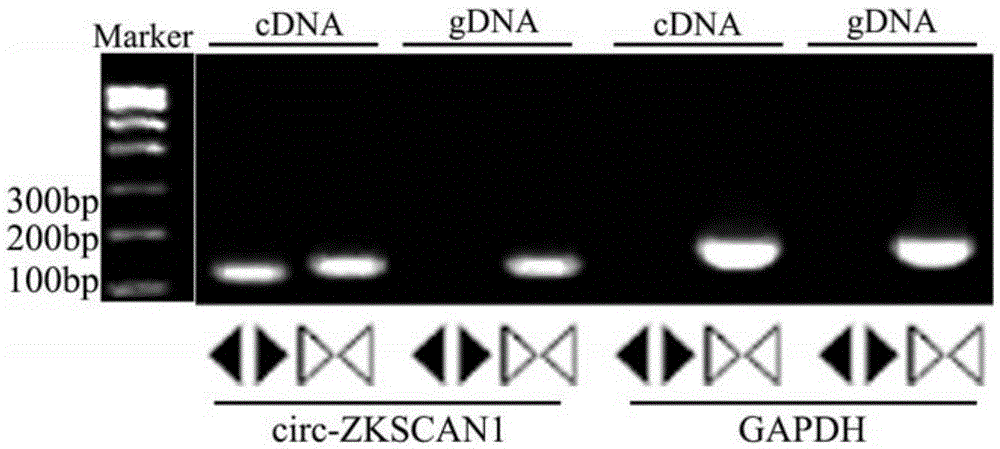
[0034] Example 1: RT-PCR reaction detection of circ-ZKSCAN1 gene expression in liver cancer tissue.
[0035] The specific experimental plan is as follows:
[0036] 1. RNA extraction
[0037] 1) Tissue processing: Take about 10 mg of tissue and add 1 ml Trizol, homogenize with a homogenizer; centrifuge for 15 minutes at 12000 g, and take the supernatant.
[0038] 2) Add 200ul chloroform to the supernatant, mix vigorously up and down for half a minute, and let stand for 3 minutes.
[0039] 3) Centrifuge at 12000g for 15 minutes at 4°C. At this time, it can be seen that the lysate is divided into three layers: the upper layer is RNA in the aqueous phase; the middle layer is DNA, lipids, etc.; the lower layer is cell residues, proteins, polysaccharides, etc.
[0040]4) Take the supernatant into a new EP tube; add an equal volume of isopropanol, mix well, let stand for 10 minutes, and then centrifuge at 12000g for 10 minutes at 4°C.
[0041] 5) Carefully remove the supernatant, ...
Embodiment 2
[0052] Example 2: Detection of the expression of circ-ZKSCAN1 in liver cancer by QPCR
[0053] 1. RNA extraction: with embodiment 1;
[0054] 2. cDNA reverse transcription: same as Example 1;
[0055] 3. QPCR amplification experiment
[0056] 1) Experimental system:
[0057]
[0058] The circ-ZKSCAN1 divergent primer of Example 1 was used to amplify the circular RNA circ-ZKSCAN1, and the hsaGAPDHconvergent primer of Example 1 was used to amplify the internal reference gene.
[0059] 2) Reaction conditions:
[0060] Step 1: 95°C for 2 minutes
[0061] Step 2 (40 cycles): 95°C for 3 seconds, 60°C for 30 seconds
[0062] The third step 60-95 ℃ melting curve
[0063] 3) Amplify the target gene on the machine
[0064] 4) qPCR relative quantitative results
[0065] The formula for calculating the relative expression of the target gene is: 2-ΔΔCt=2-[(ΔCt)Test-(ΔCt)Control]. Ct object is the Ct value of the target gene, and Ct housekeeper is the Ct value of the housekeepin...
Embodiment 3
[0068] Example 3: Construction of shcirc-ZKSCAN1 lentivirus and its stable cell line
[0069] 1. Construction of shcirc-ZKSCAN1 lentiviral vector: The shRNA sequence was synthesized in Shanghai Jierui Company, the sequence was annealed into a double-stranded DNA fragment, and inserted into the LV008 (pLL-U6-shRNA-CMV-GFP-Puro) vector through the multiple cloning site , recombinant plasmids were identified by sequencing. The nucleotide sense strand and antisense strand sequences of shcirc-ZKSCAN1 are shown in SEQ ID NO:10 and 11. The nucleotide sense strand and antisense strand sequences of the Control negative control are shown in SEQ ID NO: 12 and 13.
[0070] 2. Lentiviral packaging
[0071] (1) 24 hours before transfection, 293T cells in the logarithmic growth phase were digested with trypsin, passaged to a 10 cm cell culture dish, and cultured in a 37° C., 5% CO2 incubator. After 24 hours, when the cell density reaches 70%-80%, it can be used for transfection. Cell sta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com