A lentiviral vector expressing exosome markers and its construction method and application
A lentiviral vector and exosome technology, applied in the field of genetic engineering, can solve problems such as hindering research progress and weak brightness, and achieve the effect of strong fluorescence and easy instant observation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
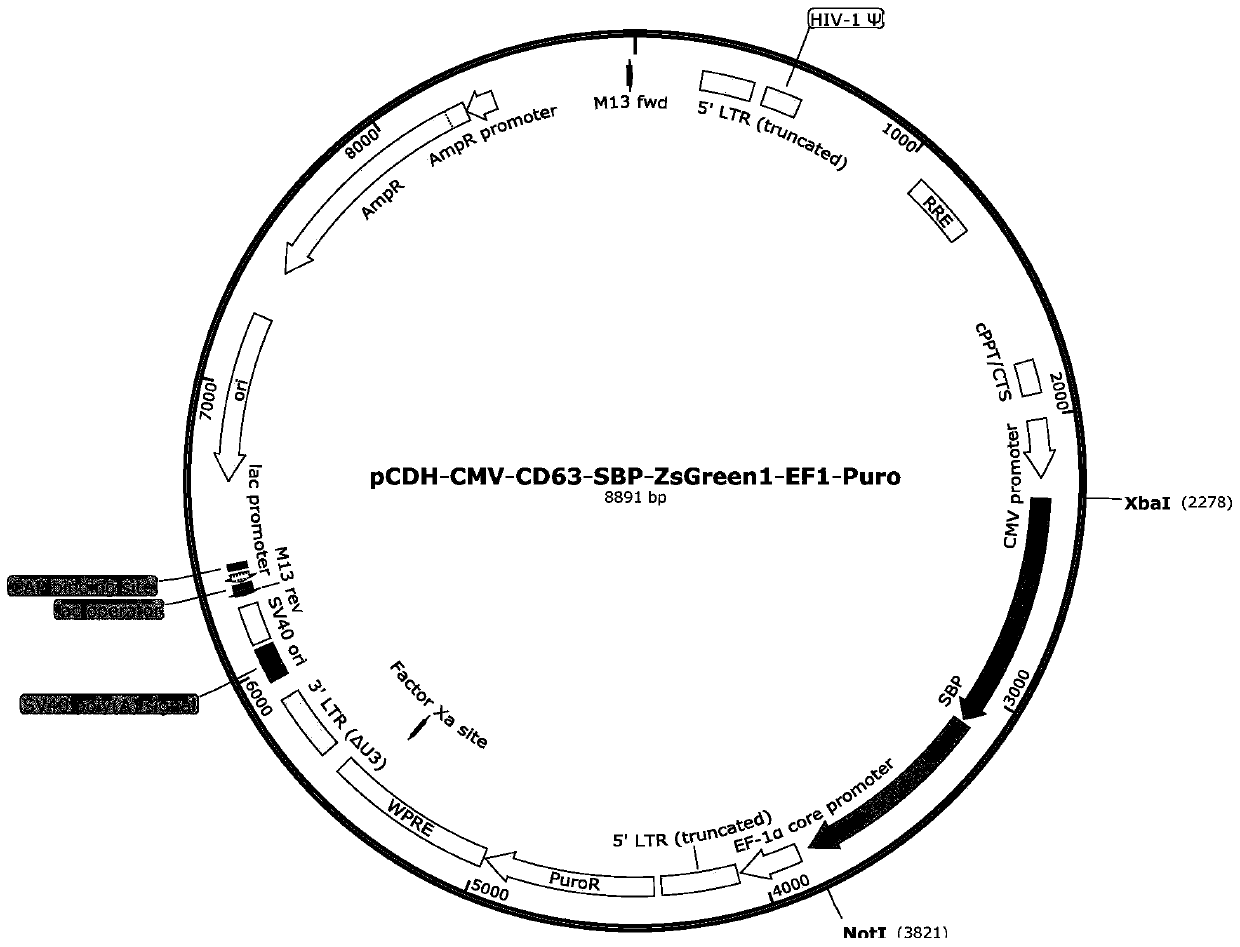
[0033] Construction of pCDH-CMV-CD63-SBP-ZsGreen1-EF1-Puro vector:
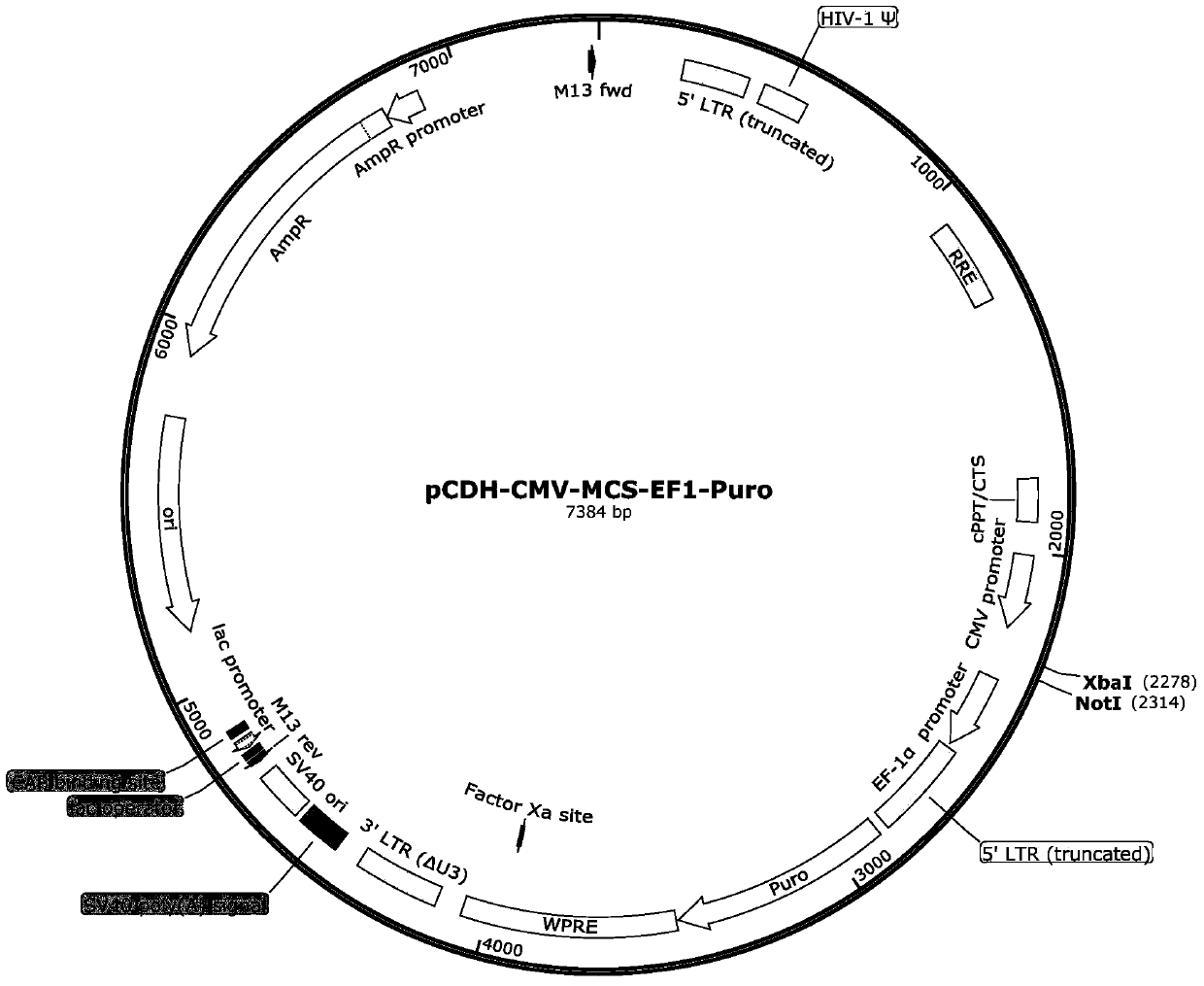
[0034] The pCDH-CMV-CD63-SBP-ZsGreen1-EF1-Puro vector was constructed on the basis of pCDH-CMV-MCS-EF1-Puro.
[0035] The pCDH-CMV-MCS-EF1-Puro vector has a CMV promoter, a multiple cloning site MCS, a selective marker Puromycin, an ampicillin resistance gene, etc., forming a 7.4kb DNA fragment. The CD63-SBP-ZsGreen1 sequence was added to the multiple cloning site between the CMV promoter and Puromycin to construct the vector pCDH-CMV-CD63-SBP-ZsGreen1-EF1-Puro.
[0036] The specific construction process of the carrier is:
[0037] 1. According to the genetic information of the CDS sequence, SBP tag sequence, and ZsGreen1 tag protein sequence on human CD63 mRNA in GeneBank, the XbaI-CD63-SBP-ZsGreen1-NotI gene DNA fragment was designed and sent to Shanghai Invitrogen Company for synthesis.
[0038] 2. Use restriction enzymes XbaI and NotI to digest the double-stranded DNA molecules synthesized in step 1 (sy...
Embodiment 2
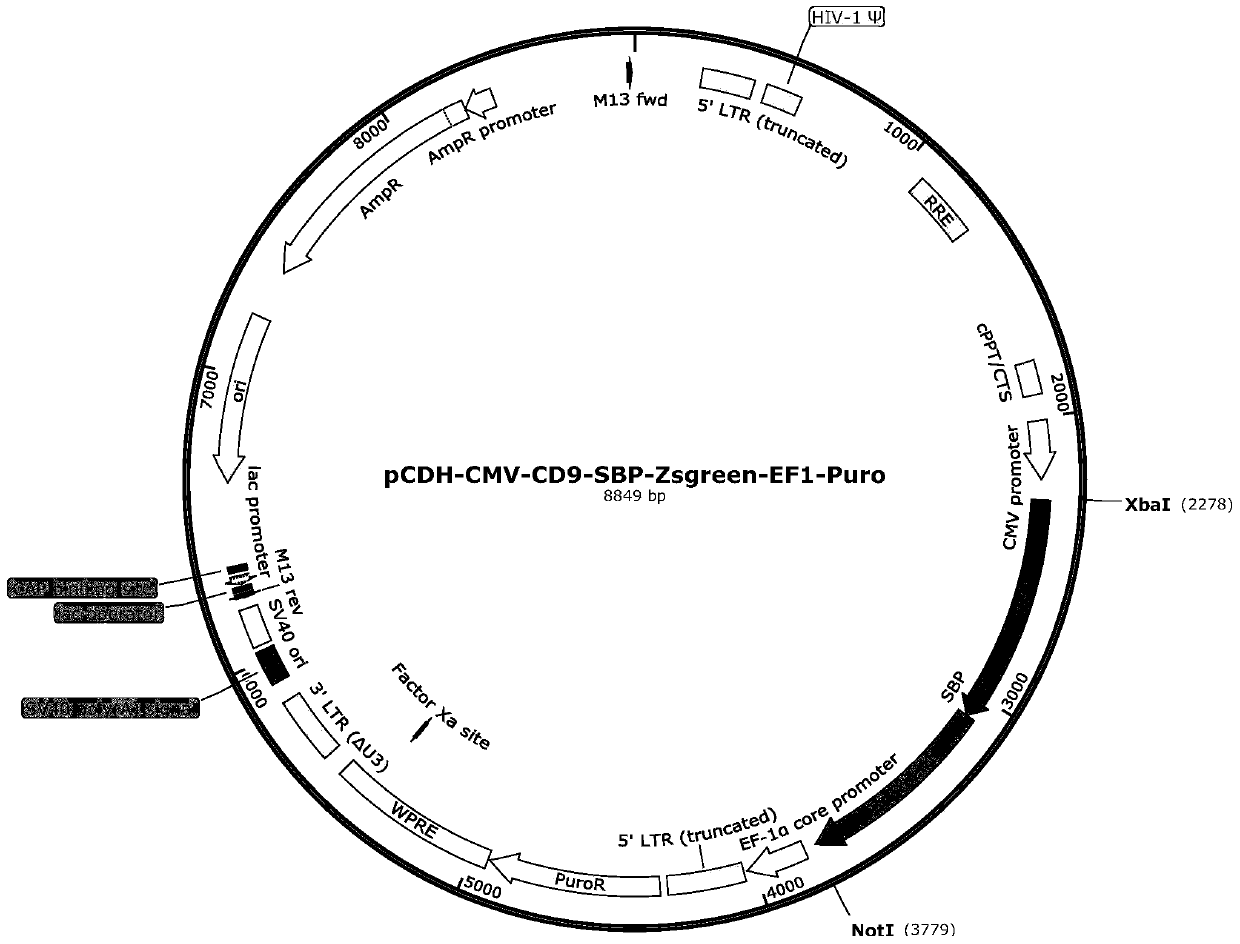
[0042] Construction of pCDH-CMV-CD9-SBP-ZsGreen1-EF1-Puro vector:
[0043] The pCDH-CMV-CD9-SBP-ZsGreen1-EF1-Puro vector was constructed on the basis of pCDH-CMV-MCS-EF1-Puro.
[0044] The pCDH-CMV-MCS-EF1-Puro vector has a CMV promoter, a multiple cloning site MCS, a selective marker Puromycin, an ampicillin resistance gene, etc., forming a 7.4kb DNA fragment. The CD9-SBP-ZsGreen1 sequence was added to the multiple cloning site between the CMV promoter and Puromycin to construct the vector pCDH-CMV-CD9-SBP-ZsGreen1-EF1-Puro.
[0045] The specific construction process of the carrier is:
[0046] 1. According to the genetic information of the CDS sequence, SBP tag sequence, and ZsGreen1 tag protein sequence on human CD9 mRNA in GeneBank, the DNA fragment of XbaI-CD9-SBP-ZsGreen1-NotI was designed and sent to Shanghai Invitrogen Company for synthesis.
[0047] 2. Use restriction enzymes XbaI and NotI to digest the double-stranded DNA molecules synthesized in step 1 (synthesize...
Embodiment 3
[0051] Application of pCDH-CMV-CD63-SBP-ZsGreen1-EF1-Puro and pCDH-CMV-CD9-SBP-ZsGreen1-EF1-Puro vectors in cell infection and cell line construction
[0052] 1. Transient transfection
[0053] (1) Cells: Take out cryopreserved HEK 293T cells from liquid nitrogen, quickly dissolve them in a 37°C water bath, then collect the cells by centrifugation, and culture them on polylysine-coated cell culture dishes after dilution with medium middle. When the density reaches 60%-70%, it can be used for subsequent transfection experiments.
[0054] (2) Transfection: 30–40 μg DNA (pCDH-CMV-CD63-SBP-ZsGreen1-EF1-Puro or pCDH-CMV-CD9-SBP-ZsGreen1-EF1-Puro vector: 15 μg without endotoxin, pLP- 1: 10 μg, pLP-2: 10 μg, pLP-VSVG: 5 μg) were transfected into the above cells. Add the DNA-calcium phosphate mixture to the cultured cells, and replace the medium after culturing for 8-12 hours. After continuing to cultivate for 60 hours, collect the cell culture medium, centrifuge at 4000g for 5 min...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com