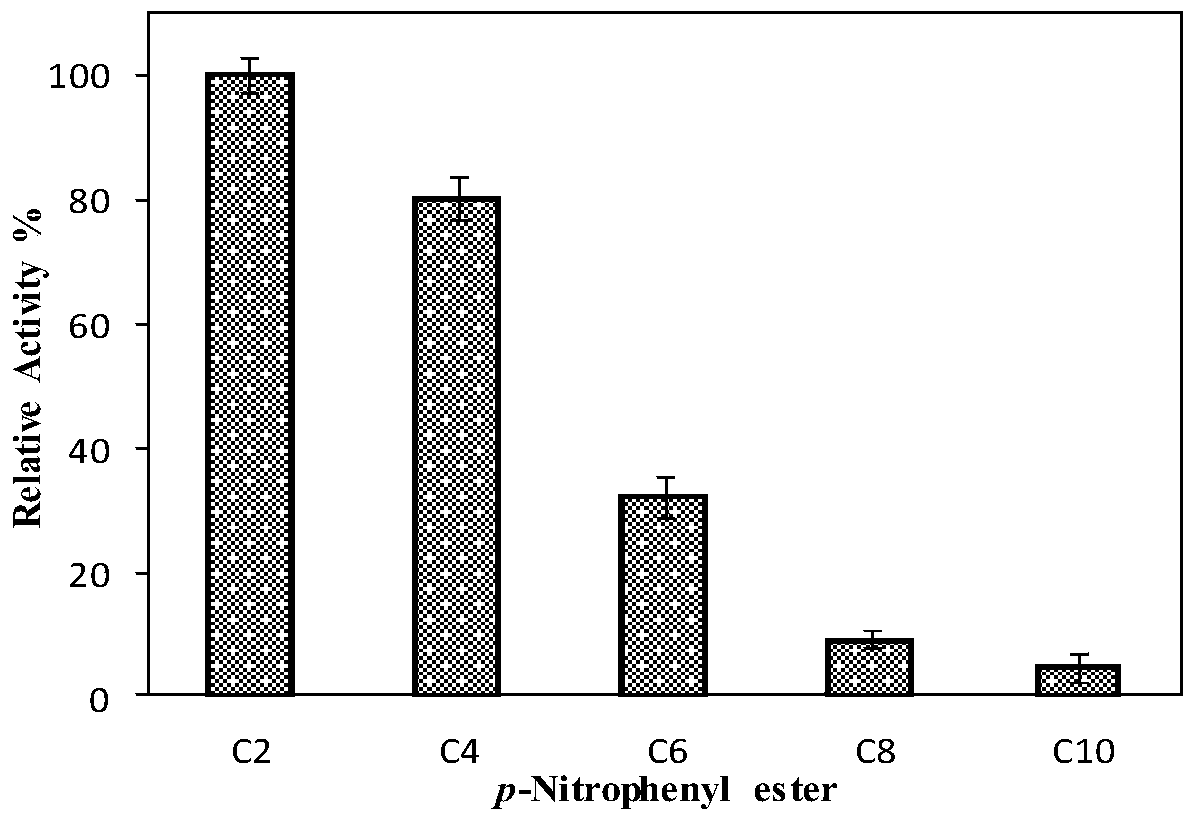
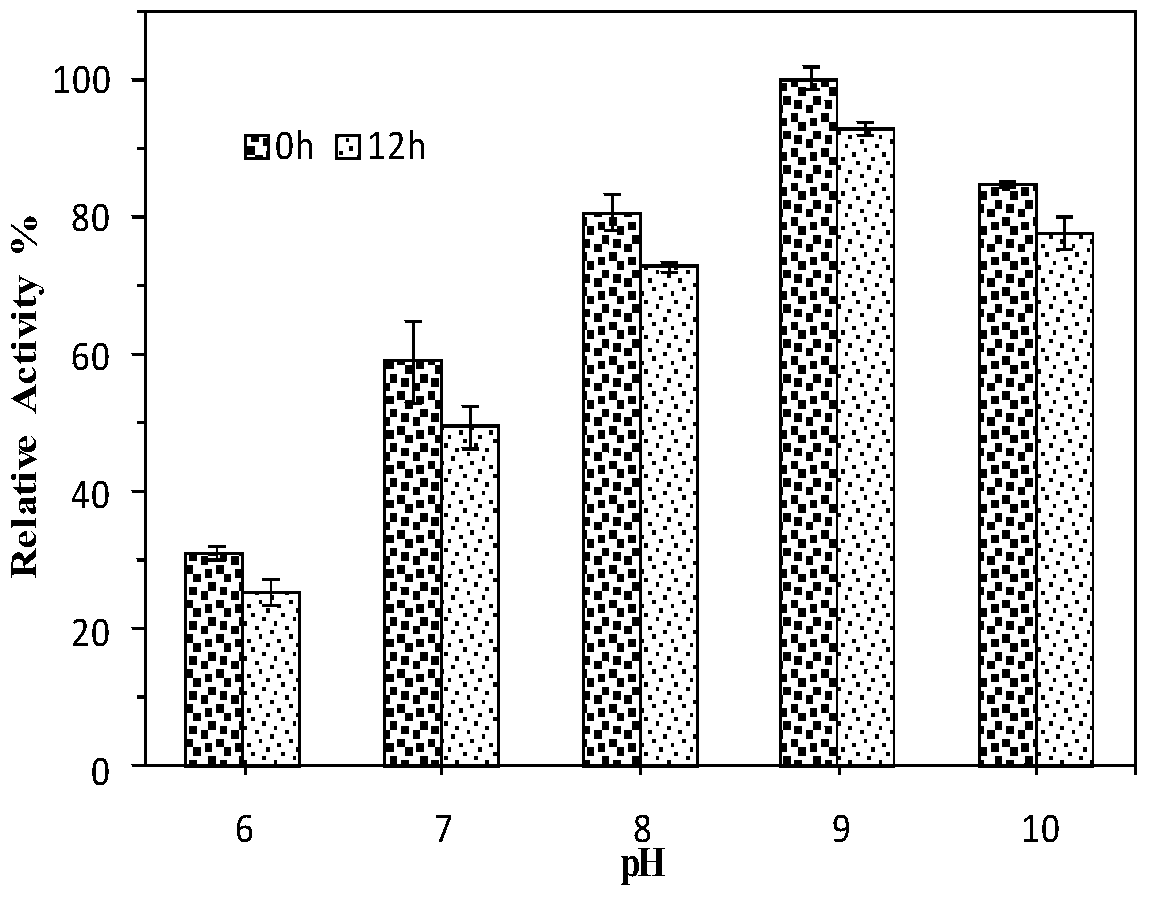
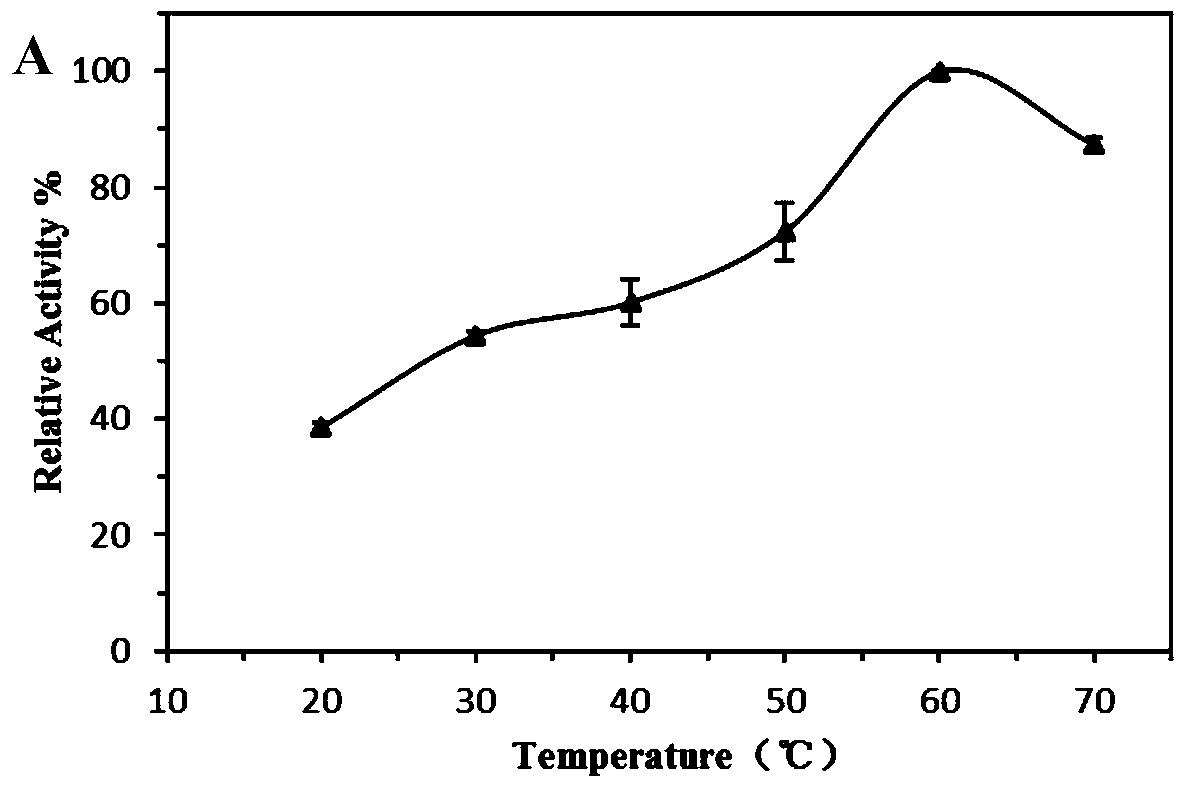
A kind of esterase phe14 and its coding gene and application
A technology of PHE14 and esterase, applied in application, genetic engineering, plant gene improvement, etc., can solve problems such as expensive esterase and production technology constraints
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: Design of primers for esterase gene PHE14 and determination of open reading frame boundaries
[0030] Genomic DNA of Pseudomonadaceae oryzihabitans HUP022 was extracted, and after sequencing and verification, the genome was annotated by means of bioinformatics, the esterase gene was analyzed, and the open reading frame of the esterase gene PHE14 was determined. The nucleotide sequence is shown in SEQ ID NO.1, the full length is 645bp (from the start codon to the stop codon), and the amino acid sequence of the esterase PHE14 encoded by it is shown in SEQ ID NO.2, a total of 214 Amino acid, the gene is a brand new esterase gene. According to the sequence of the esterase gene PHE14 obtained from the analysis, the primers were designed as follows: Forward primer: 5′-CAC GAATTC GTGCTGGAATCGCCTAGC-3′, the underlined part is the EcoRI restriction site; reverse primer: 5′-CCG CTCGAG TTATTTTTTGCCGAGACGTGCC 3′, the underlined part is the Xho I restriction site.
Embodiment 2
[0031] Embodiment 2: Cloning and vector construction of esterase gene PHE14
[0032] 2.1PCR amplification
[0033] The primer (forward primer: 5'-CAC GAATTC GTGCTGGAATCGCCTAGC-3′, reverse primer: 5′-CCG CTCGAG TTATTTTTTGCCGAGACGTGCC-3′) was sent to Shanghai Bioengineering Co., Ltd. to synthesize primers. The synthesized primers were diluted to 10 μM with TE, and the total DNA of Pseudomonadaceae oryzihabitans HUP022 extracted was used as a DNA template to establish the reaction shown in Table 1. system:
[0034] Table 1 PCR reaction system
[0035]
[0036] Amplify the esterase gene PHE14 using the following PCR amplification program: a. Denaturation at 94°C for 3 minutes; b. Denaturation at 94°C for 30 s, annealing at 55-65°C for 0.5-1 min, extension at 72°C for 1 min, and 20 cycles; c. 72°C Extend for 10 min and cool to 10°C.
[0037]The PCR product was electrophoresed in 1% agarose gel for 20 min at 120V, and observed in a gel imaging system. The band around 645b...
Embodiment 3
[0047] Embodiment 3: High expression of esterase gene PHE14 in Escherichia coli BL21 (DE3)
[0048] 3.1 Preparation of Escherichia coli BL21(DE3) Competent Cells
[0049] 1. Inject a small amount of Escherichia coli BL21(DE3) into a 5mL LB test tube, shake overnight at 37°C, 250rpm;
[0050] 2. Inoculate the Escherichia coli BL21 (DE3) bacterial solution after overnight shaking culture into a 300ml LB shake flask at an inoculum size of 1% volume ratio, and shake it at 37°C for 3-4h (≥ 300rpm) to obtain the original culture;
[0051] 3. Rapidly cool the cultured shake flask to 0°C in ice water, divide the original culture into ice-precooled centrifuge tubes (50 mL), and place on ice for several minutes;
[0052] 4. Centrifuge at 4000rpm for 10min at 4°C to recover the cells and remove the supernatant;
[0053] 5. Pre-cooled with ice 10mL 0.1M CaCl 2 Resuspend the cells and centrifuge at 4000rpm for 10min at 4°C to recover the cells;
[0054] 6. Repeat 5, use 10mL 0.1M CaCl ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com