Method for achieving bacillus licheniformis gene knockout rapidly
A Bacillus licheniformis, gene knockout technology, applied in biochemical equipment and methods, glycosylase, virus/bacteriophage, etc., can solve the problems of complex construction process, complex carrier structure, long operation cycle, etc., and achieve the construction process Simple, efficient integration process, and short gene knockout cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
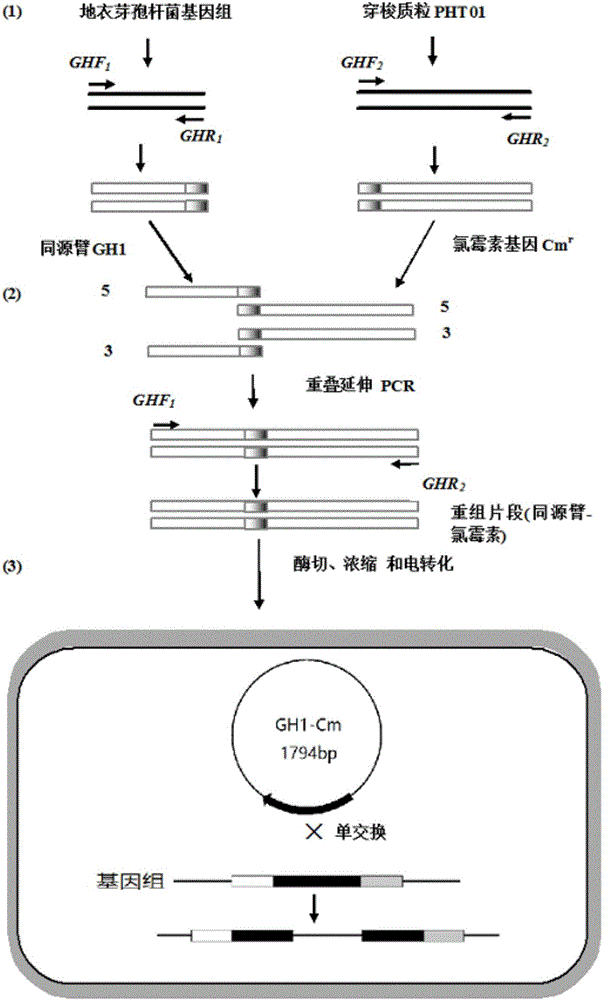
[0076] Gene knockout fragment construction
[0077] (i) extracting the DNA of the Bacillus licheniformis thallus, using the DNA as a template, performing PCR amplification to obtain the homology arm GH1;
[0078] Described PCR primer sequence is as follows:
[0079] f 1 :CG GGATCC AACCGTTTCTTTTATCCGCAGT
[0080] R 1 :CCAGCAAAATTCAGCATCTCTCGGTTAAGCA
[0081] Among them, the underline marks the BamHI restriction site;
[0082] The PCR amplification system is 50 μl:
[0083] 2×HiFi-PCRmaster25μl, 10μmol / L primer F12.0μl, 10μmol / L primer R12.0μl, template 2.0μl, use ddH 2 O make up 50 μl;
[0084] The PCR amplification procedure is as follows:
[0085] Pre-denaturation at 95°C for 5 min; denaturation at 94°C for 30 sec, annealing at 57°C for 30 sec, extension at 72°C for 1.5 min, 30 cycles; extension at 72°C for 10 min, storage at 4°C;
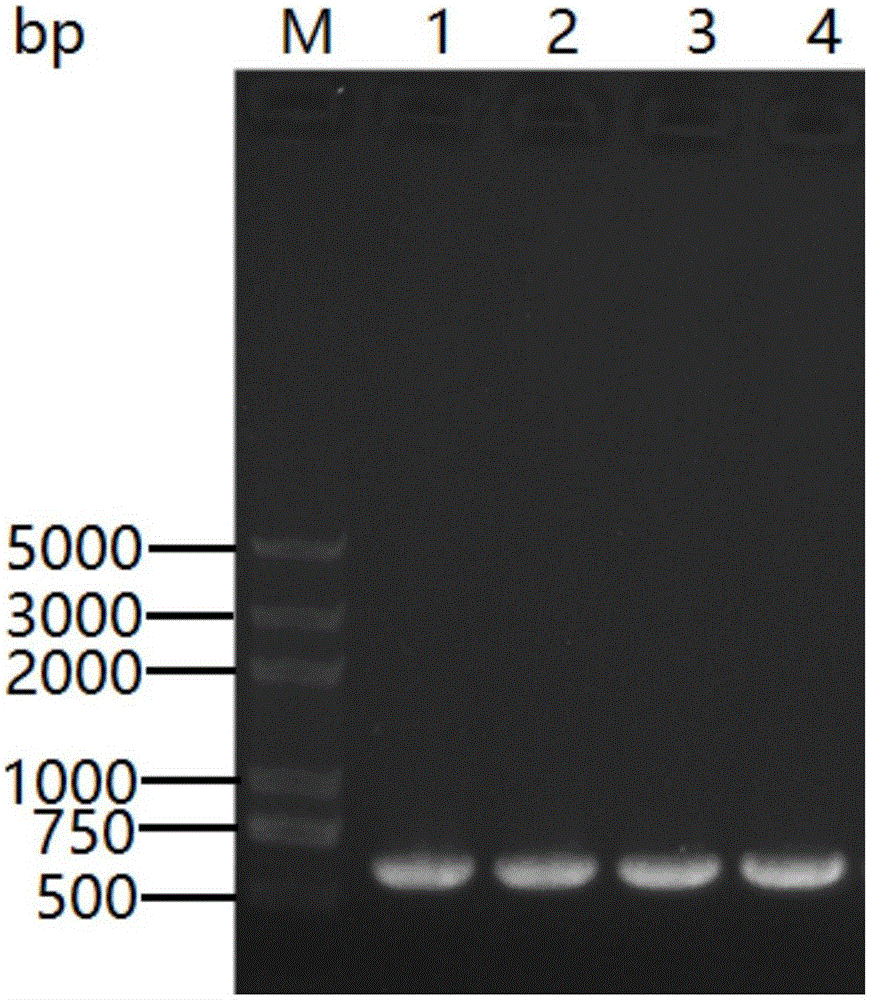
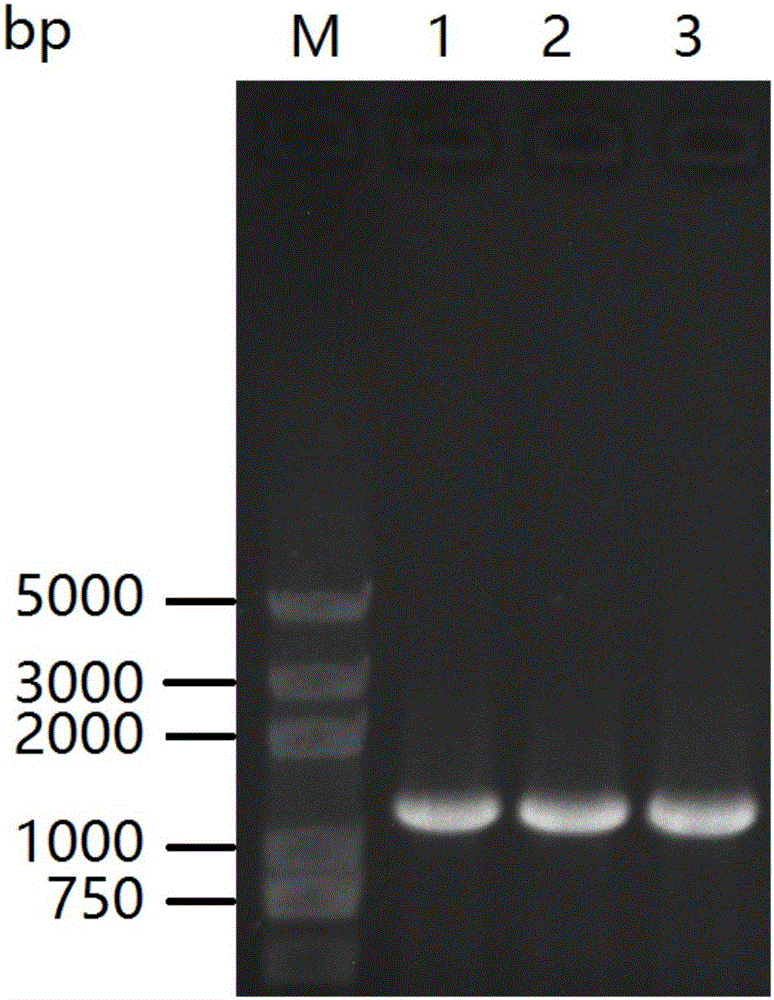
[0086] Agarose gel electrophoresis test PCR product, length is 549bp (SEQ ID NO.3, such as figure 2 shown), use the SanPrep Column DN...
Embodiment 2
[0109] Preparation of Bacillus licheniformis competent
[0110] (i) Pick a single colony of Bacillus licheniformis on the surface of fresh LB solid medium, inoculate it in 10mL of GM (expansion) medium, cultivate overnight at 37°C and 220r / min;
[0111] (ii) Take 1mL of the above bacterial solution and transfer it to 100mL of GM (expansion) liquid medium, and cultivate it to OD at 37°C and 220r / min 600 =0.9;
[0112] (iii) Transfer the bacterial solution to a 100mL centrifuge tube and place in an ice bath for 20 minutes to stop the growth of the bacterial cells;
[0113] (iv) Centrifuge at 4°C, 5000g, 5min after ice bath, and collect the bacteria;
[0114] (v) The thalline after centrifugation is washed 3 times with pre-cooled electroporation buffer (ETM);
[0115] (vi) After washing, use 1000uL electroporation buffer to resuspend the bacteria;
[0116] (vii) Aliquot the prepared competent cells into 100 μL tubes and store at -80°C for later use.
[0117] Among them, GM: ...
Embodiment 3
[0120] GH1-Cm r Fragment transformation of Bacillus licheniformis cells
[0121] (i) the GH1-Cm that embodiment 1 makes r The fragment was digested with restriction endonuclease BamHI;
[0122] Enzyme digestion system (40uL) is as follows:
[0123]
[0124] (ii) Concentrate and purify the digested product
[0125] (1) Add 1 / 10 volume of 3M sodium acetate and 2.5 volumes of absolute ethanol, and place in a -20°C refrigerator for 20 minutes;
[0126] (2) Centrifuge at 12000r / min for 5min to obtain a precipitate;
[0127] (3) 300 μL of 75% ethanol by volume to resuspend the pellet;
[0128] (4) Centrifuge at 12000r / min for 5min, remove ethanol, and air-dry at 37°C for 30min;
[0129] (5) Add 15-18μL ddH 2 O Resuspend DNA and store at -20°C.
[0130] (iii) Electroconversion
[0131] Determination of GH1-Cm by Nucleic Acid Ultramicro Spectrophotometer r Fragment concentration, after reaching a concentration of 2000μg / ml, perform electroporation, the electroporation con...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com