Prediction method for protein post-translational modification methylation loci
A post-translational modification and prediction method technology, applied in the field of protein post-translational modification methylation site prediction, can solve problems such as single features, over-modeled, and lack of detailed classification, achieving high throughput and high accuracy Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
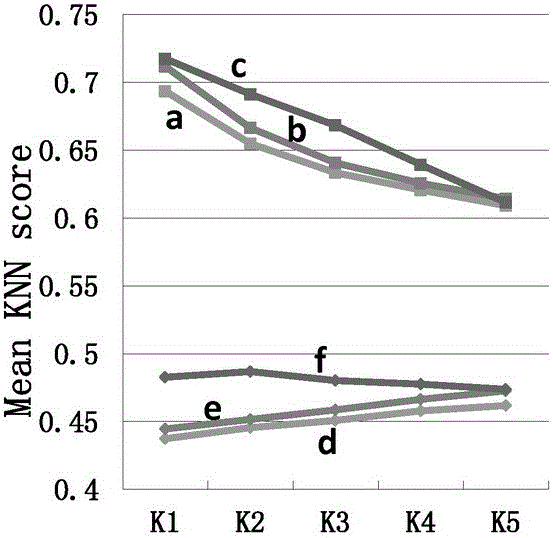
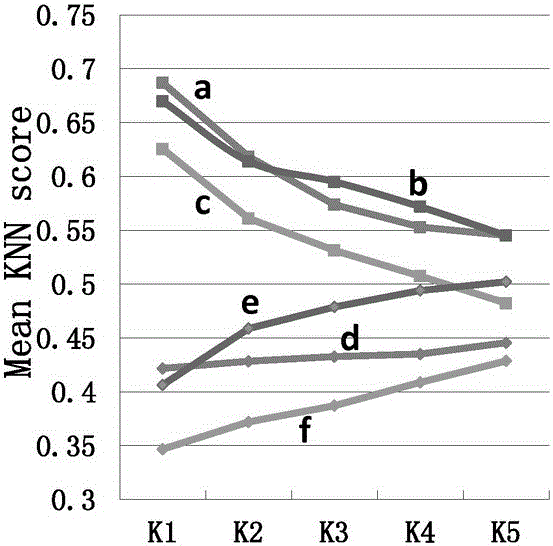
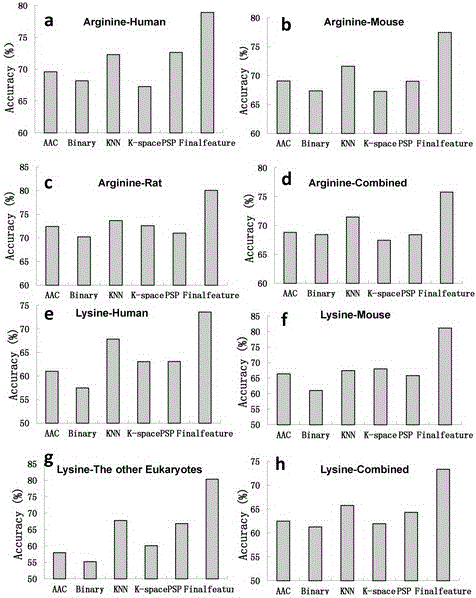
[0036] The protein methylation data are collected from protein databases such as UniProt and PhosphoSite. The positive samples are the methylation sites marked by experimental verification, and the negative samples are randomly selected from the same protein as the positive samples. Labeled arginine (R) and lysine (K) sequences. The protein sequences collected above were removed 30% homology by the cd-hit tool, and then uniformly cut into a sequence with R or K as the center, 9 amino acids upstream, 9 amino acids downstream, and a length of 19. The sequence information, evolution information and physical and chemical properties of the positive and negative sample sequences after unified cutting and preprocessing are encoded according to the following steps:
[0037] (1) Sequence information encoding of sample sequence: sequence information includes amino acid occurrence frequency, binary code and K-space amino acid pair; Convert each amino acid in the sequence into a 20-dimen...
Embodiment 2
[0057] In order to facilitate the prediction application of protein methylation sites, an online prediction platform (http: / / bioinfo.ncu.edu.cn / PSSMe.aspx) was developed based on PSSMe and combined programming with MATLAB and C#. Just enter the protein name or fasta format sequence of the protein to be predicted in the UniProt database in the designated area of the website, and the possible methylation site prediction of the protein can be performed. For example, if the user wants to predict the methylation site of the protein sequence named "B4DEH8", just enter "B4DEH8" in the protein name of the website, click the "Load" button, and the PSSMe tool will automatically download the protein from the UniProt database sequence and import it into the designated area, the B4DEH8 protein sequence information is as follows:
[0058] >tr|B4DEH8|B4DEH8_HUMAN
[0059] MEEEAEKLKELQNEVEKQMNMSPPPGNAGPVIMSIEEKMEADARSIYVGNVDYGATAEELEAHFHGCGSVNRVTILCDKFSGHPKGFAYIEFSDKESVRTSLALDESLFRGRQIKVIP...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com