Gene chip for detecting beta-proteobacteria communities in marine environment
A technology of gene chip and chip carrier, applied in the field of gene chip for detecting gamma-proteobacteria community in marine environment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
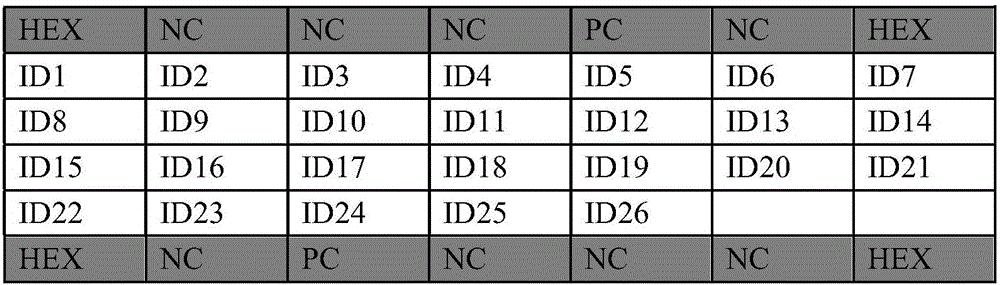
[0024] Example 1: Design of probes for gene chips for the detection of β-proteobacterial communities
[0025] The community information of β-proteobacteria in seawater of East China Sea was obtained by high-throughput sequencing technology and clone library technology. According to the 16S rRNA gene sequence information of the β-proteobacteria community, probes were designed for 31 major β-proteobacteriaceae (Alcaligenaceae, Burkholderiaceae, Comamonadaceae, Hydrogenophilaceae, Methylophilaceae, Neisseriaceae, Nitrosomonadaceae, Oxalobacteraceae, Rhodocyclaceae) in the seawater of the East China Sea, and Verify in BLAST to obtain the probe sequence used to prepare the gene chip.
[0026] From the perspective of detection specificity, the applicant screened the above-mentioned sequences that may be used as probes, and finally determined the following probe sequences:
[0027] Wherein the nucleic acid probe for detecting Alcaligenaceae microorganisms has a nucleotide sequence o...
Embodiment 2
[0040] Embodiment 2: the specific detection of the β-proteobacteria colony detection gene chip of the present invention
[0041] The representative clone UnculturalComamonadaceae bacterium clone 2-8 of the seawater dominant family Comamonadaceae in β-proteobacteria was selected to detect the specificity of the gene chip for β-proteobacteria community detection.
[0042] ① DNA extraction: Plasmid DNA was extracted with a plasmid extraction kit.
[0043] ② 16S rRNA gene amplification: The 16S rRNA gene was linearly amplified using the cloning vector pEASY-T1 universal primers M13F (GTA AAA CGACGG CCA GT) and M13R (GTC CTT TGT CGA TAC TG). The amplification program was 94° C. for 5 min, 25 cycles (94° C. for 30 s, 55° C. for 30 s, 72° C. for 1 min and 30 s), and 72° C. for 10 min.
[0044] ③ Fluorescent labeling: Use fluorescently labeled random primers (Cy3-NNN NNN NNN) and Klenow enzyme to label the 16SrRNA gene amplification product. 1.5h at 37°C, 10min at 70°C.
[0045] ④ ...
Embodiment 3
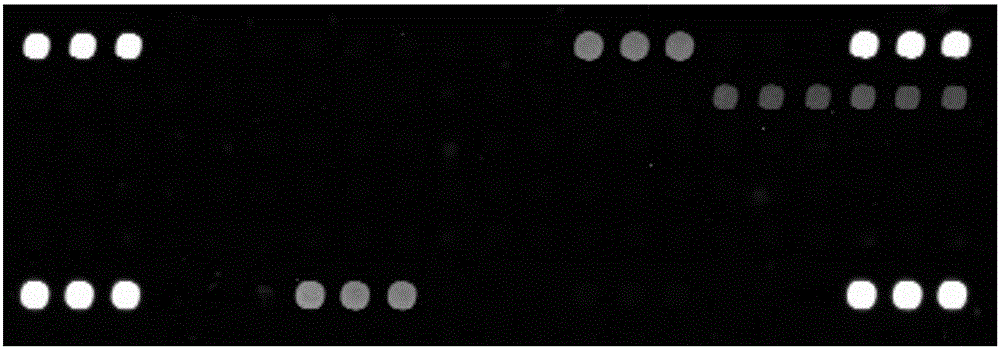
[0047] Example 3: Detection of β-proteobacteria species in seawater samples using chips
[0048] The seawater sample 22A was hybridized with the marine β-proteobacteria community detection gene chip of the present invention, the specific method is as in Example 2. And compared with the results of high-throughput sequencing. The results are shown in Table 1.
[0049] Table 1 Detection results of β-proteobacteria in seawater sample 22A
[0050]
[0051] The gene chip detection results showed that the β-proteobacteria contained in seawater sample 22A included Burkholderiaceae, Comamonadaceae, Methylophilaceae, Nitrosomonadaceae, Oxalobacteraceae; the high-throughput sequencing results showed that seawater sample 22A contained β-proteobacteria including Comamonadaceae, Methylophilaceae, Neisseriaceae, Nitrosomonadaceae, Rhodocyclaceae. The detection result of the gene chip of the present invention is generally consistent with the result of high-throughput sequencing, indicat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com