DNA pulldown method and kit
A technology of reagents and magnetic beads, applied in the biological field, can solve the problems of the influence of nuclease contamination and the low specificity of DNA probe and protein binding, and achieve the effects of preventing contamination, saving experimental costs and improving specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
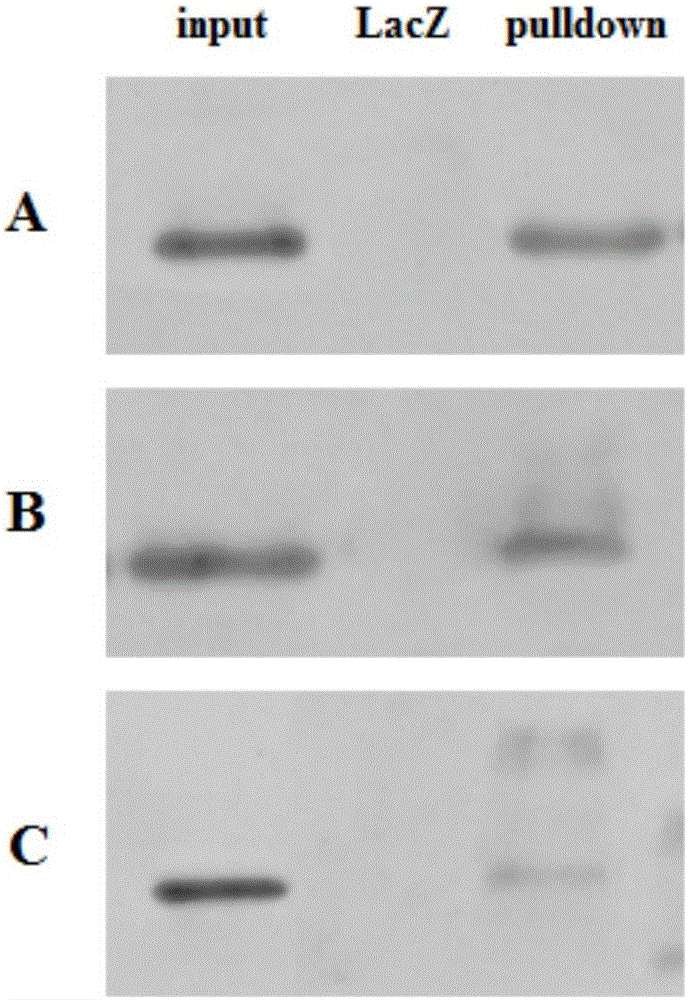
[0038] In this example, specific DNA probes were designed for the P53 protein in the target region, and the probes were labeled with desthiobiotin 8-oxoG. The design and labeling methods of probes are well known to those skilled in the art and will not be repeated here. Streptavidin coupled to magnetic beads (BeaverBeads TM Streptavidin, Beaver Biotechnology Co., Ltd.) and affinity binding with desthiobiotin; nuclei extracts were incubated with magnetic beads-DNA probes, and the protein molecules could specifically bind to the probes; after washing, the non-specific binding proteins could be Molecule removal; finally, elution is performed with an eluent (for streptavidin) to obtain the target probe-protein complex, and then the protein type is identified by Western Blot. Specific steps are as follows:
[0039] S1. Magnetic bead pretreatment
[0040] ①Put 80 μL of streptavidin-labeled magnetic beads into an EP tube, add 1 mL of 1×TES to wash the magnetic beads;
[0041] ② ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com