Positioning method of polypeptide disulfide bond
A positioning method and disulfide bond technology, applied in measuring devices, material analysis through electromagnetic means, instruments, etc., can solve the problems of long detection cycle, low accuracy, and low throughput, and achieve long detection cycle and high cost long effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1. Simple and quick alkylation of partial reducing agent and subsequent total reduction
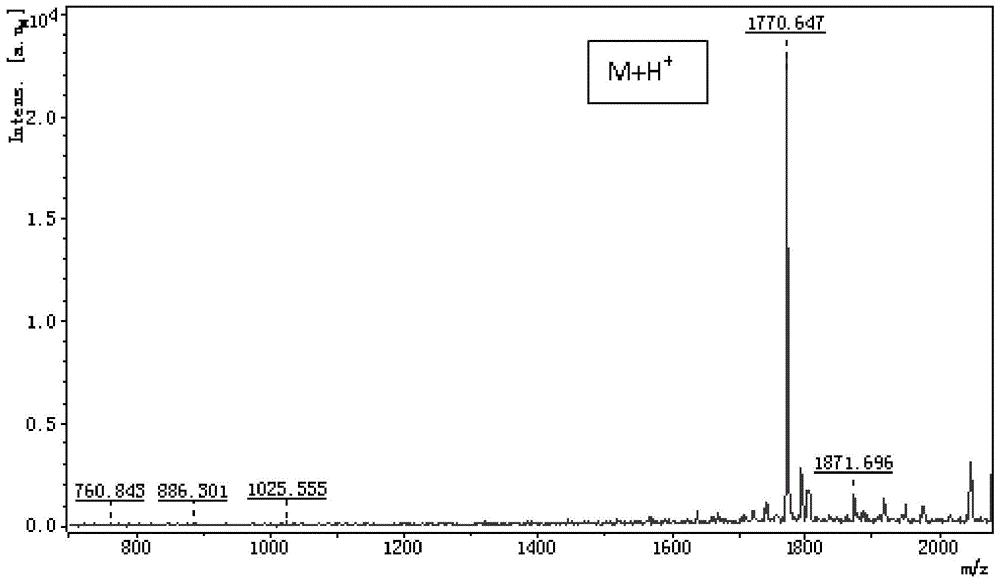
[0081] In this embodiment, a conotoxin polypeptide sample containing three pairs of disulfide bonds (molecular weight 1769.69Da, sequence: RCCVHPACHDCICCIT, MALDI-TOF detection mass spectrum is as follows figure 1 ) as an example to illustrate the positioning method of polypeptide disulfide bonds.
[0082] 1. Partial restoration
[0083] Weigh 0.1mg of conotoxin polypeptide sample containing three pairs of disulfide bonds, dissolve in 1ml ddH 2 In O, DTT (sigma) was added so that the final concentration was 10 mM, and the partial reduction was performed at room temperature for 10 min. After the reaction, use a C18 extraction column (strata TM -X, phenomenonex) to remove the remaining DTT (to avoid the disulfide bond being continuously reduced during subsequent alkylation), and the eluate was freeze-dried;
[0084] 2. Alkylation of IAM
[0085] The freeze-dried sample w...
Embodiment 2
[0088] Example 2. MALDI-TOF detection of partial reductive alkylation effect
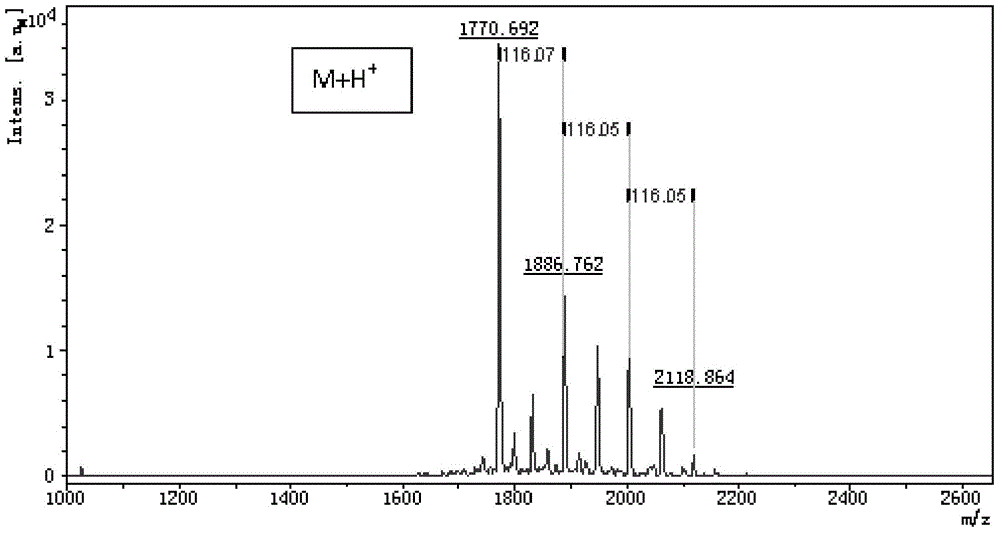
[0089] Take 1 μL of the sample after partial reductive alkylation and subsequent complete reduction, and spot it on the MALDI-TOF (Bruker) sample target. After the sample is dried, add 0.3 μL matrix (CHCA, English full name α-Cyano-4-hydroxy-cinnamic acid, the full name in Chinese is α-cyano-4-hydroxycinnamic acid), after drying, add 2.5μL 0.1% TFA aqueous solution, let it stand for 5 seconds and then quickly suck it away, so as to play the role of simple desalination. Then put the sample target into the mass spectrometer, select the primary mass spectrometry reflectance mode detection method to detect the effect of partial reductive alkylation, the results are as follows figure 2 As shown, the mass spectrum peaks corresponding to one pair, two pairs and three pairs of disulfide bonds respectively appear obviously, and the effect of partial reductive alkylation is better.
Embodiment 3
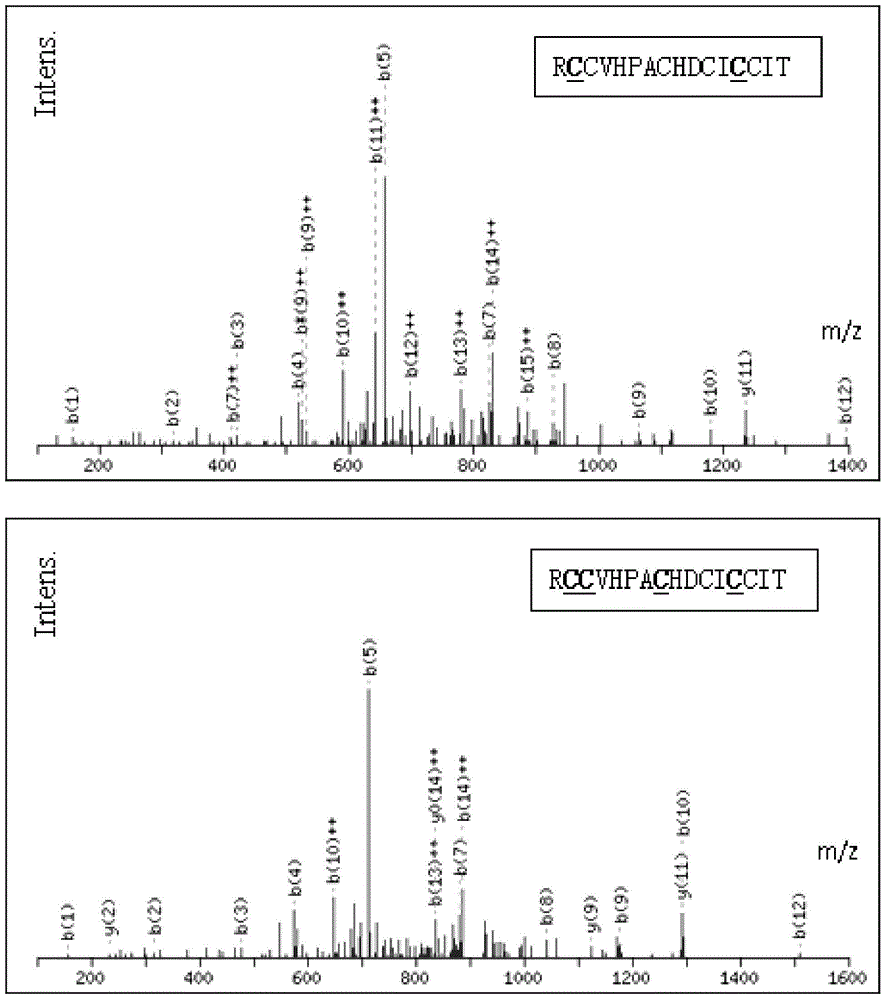
[0090] Embodiment three, LC-MS / MS mass spectrometry detection
[0091] LC-MS / MS (using ThermoFisher Scientific's Q-Exactive mass spectrometer) mass spectrometry detection of partially reduced alkylated polypeptide samples that have passed the MALDI-TOF mass spectrometry detection, the total detection time is 65min, and the core mass spectrometry parameters are as follows:
[0092] MS resolution: 70,000; scan range: 350-1800m / z; resolution of MS: 7,500; data dependent acquisition is the top 20 parent ions of MS for MS acquisition; fragmentation energy of MS is 27%; the dynamic exclusion time is set to 8 seconds;
PUM
| Property | Measurement | Unit |
|---|---|---|
| Concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com