Test method of reaction of EPHX1 genotype based COPD to NAC
A test method and genotype technology, applied in the field of testing the response of EPHX1 genotype COPD to NAC, can solve the problem of not being able to show the different curative effects of N-acetylcysteine antioxidant therapy in COPD patients
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0020] In order to make the object, technical solution and advantages of the present invention more clear, the present invention will be further described in detail below in conjunction with the examples. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
[0021] The application principle of the present invention will be described in detail below in conjunction with the accompanying drawings.
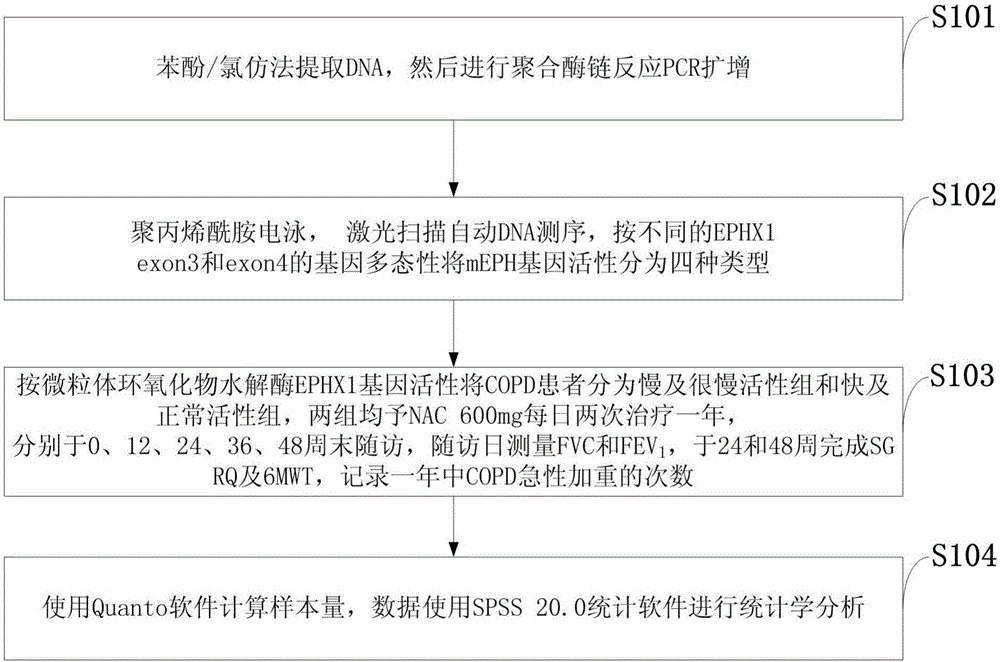
[0022] Such as figure 1 As shown, the test method based on the EPHX1 genotype COPD response to NAC in the embodiment of the present invention comprises the following steps:
[0023] S101: Extract DNA by phenol / chloroform method, and then carry out PCR amplification by polymerase chain reaction;
[0024] S102: Polyacrylamide electrophoresis, laser scanning automatic DNA sequencing, mEPH gene activity is divided into four types according to different gene polymorphisms of EPHX1exon3 an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com