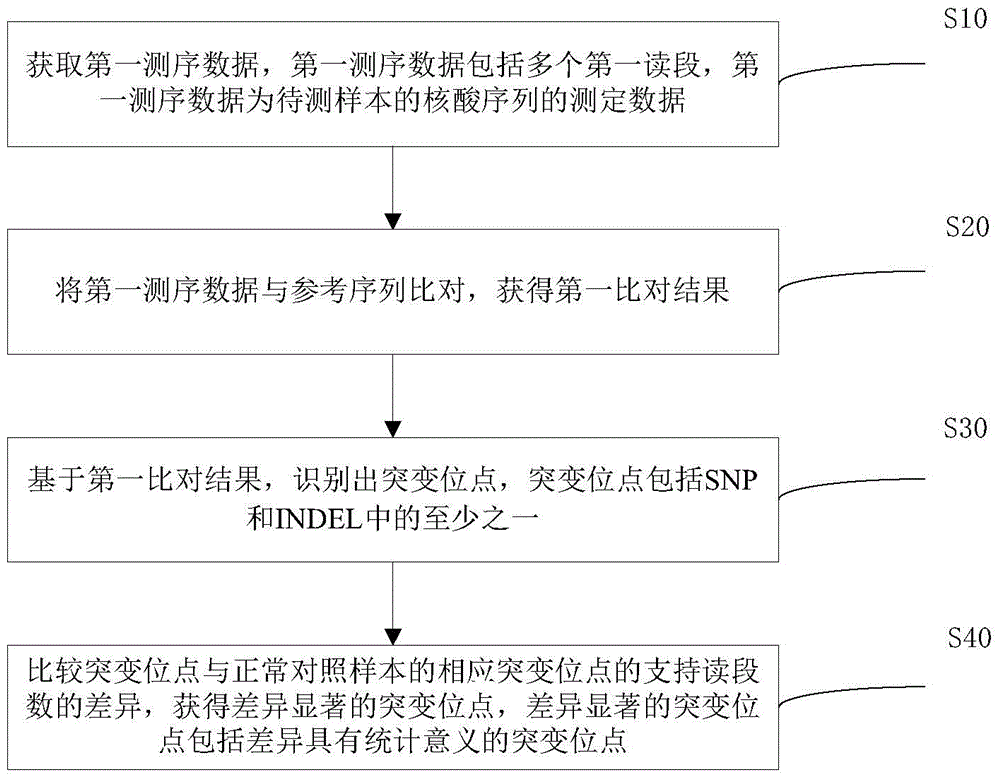
Method and apparatus for detecting mutation of somatic cells
A technology for detecting bodies and cells, applied in the field of bioinformatics, to achieve the effects of low cost, convenient transplant operation, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
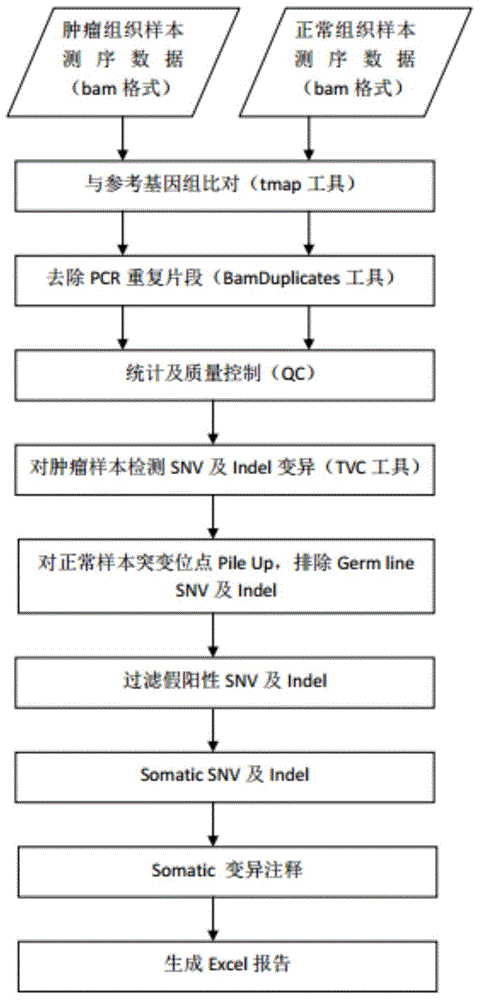
[0039] After obtaining the off-machine data of BGISEQ-100 sequencing, such as image 3 As shown, generally include the following steps:
[0040] 1. Alignment with the reference genome
[0041] Use the tmap tool to align the sequencing data to the reference genome to obtain accurate alignment results. The tmap tool comes from: https: / / github.com / iontorrent / TS / tree / master / Analysis / TMAP
[0042] 2. Remove PCR duplicates in the comparison results
[0043] The BamDuplicates tool was used to remove PCR duplicates from the results (bam format) compared with the tmap tool. Among them, the BamDuplicates tool comes from Ion Torrent Systems, Inc.
[0044] 3. Statistics and quality control
[0045] Count the ratio of the data volume of the target area to the total data volume, the average sequencing depth of the target area, the coverage rate of the target area, etc., and generate a series of quality control indicators to judge the quality of the sequencing data.
[0046] 4. Somatic...
Embodiment 2
[0075] The cancer tissue sample and blood cell sample (sample name: TJ0002) of a female patient with colorectal cancer were obtained from the hospital for target region capture and sequencing on the BGISEQ-100 platform. BamDuplicates deduplication, quality control (QC), Somatic mutation detection, mutation annotation, report generation steps, and finally obtain the patient's individualized tumor detection report.
[0076] The process method included in the first embodiment is integrated into the software Oseq-T, and the operating environment of the software is a Unix / Linux operating system, which runs through the Unix / Linux command line.
[0077] The specific operation steps are as follows:
[0078] Enter the following command in the LINUX operating system computer terminal to invoke the software:
[0079] perl oseq_chip_proton.pl –c cancer.list –n normol.list,
[0080] Oseq-T command line parameters are shown in Table 1 parameter description.
[0081] Table 1
[0082] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com