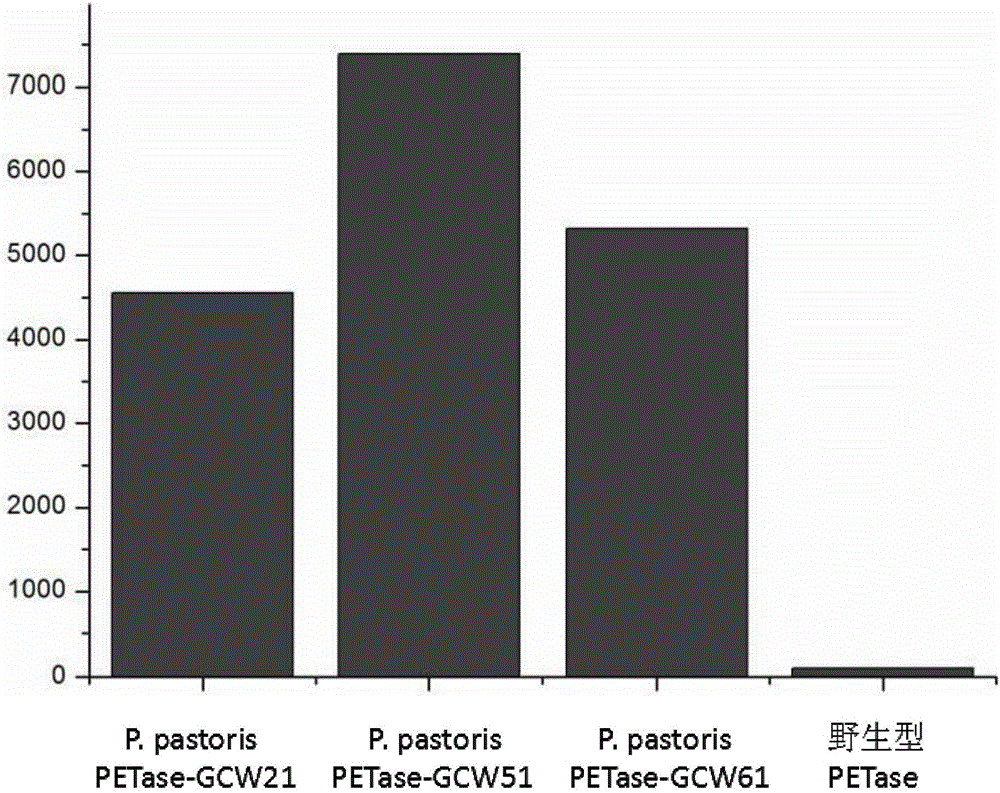
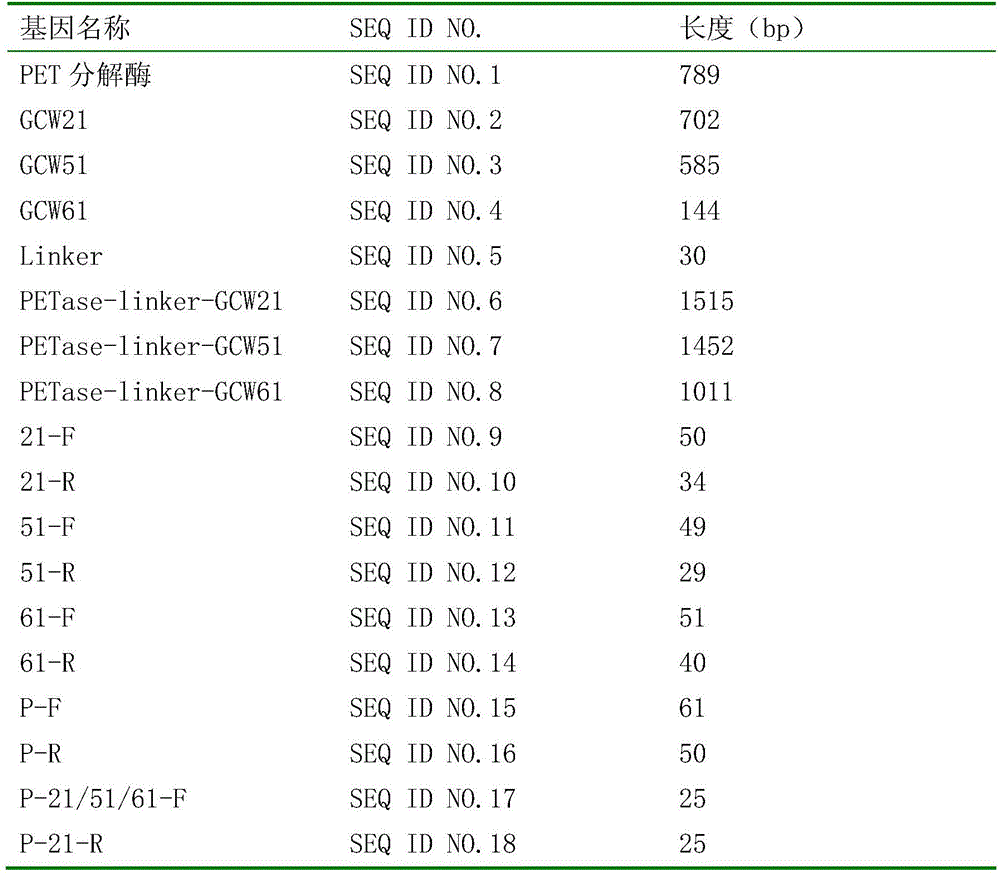
Cell surface exhibition PET decomposing enzyme reorganizable pichia pastoris and structure and application
A Pichia pastoris, cell surface technology, applied in the field of genetic engineering of enzymes, can solve the problems of unfavorable large-scale industrial application, low decomposition efficiency, difficult separation and purification, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 The construction and application of recombinant Pichia pastoris displaying PET decomposing enzyme on the cell surface, including the following steps:
[0031] chemically synthesized the nucleotide sequence of the improved PET decomposing enzyme shown in SEQ ID No.1;
[0032] Design upstream primer P-F and downstream primer P-R according to the sequence of SEQ ID No.1;
[0033] P-F: 5'-3'CCGCTCGAGAAAAGAGATTACAAGGATGACGACGATAAGCAAAACCAATCCTTATGCCCGTG
[0034] P-R: 5'-3'ACTACCACCTCTCTCCACTACCTCCACCACCGCTACAGTTGGCGGTACGAA
[0035] Perform PCR, and perform 1% agarose gel electrophoresis on the PCR reaction product to recover the target gene fragment to obtain the PET decomposing enzyme gene.
[0036] According to GCW21 (NCBI Reference Sequence: XM_002491407), after predicting its signal peptide, design upstream primer 21-F and downstream primer 21-R;
[0037] 21-F: 5'-3'GGTGGTGGAGGTAGTGGAGGAGGTGGTAGTGACCAGCGAATCTCTGTCAC
[0038] 21-R: 5'-3'CCGGAATTCTTATAACAAAGC...
Embodiment 2
[0110] Example 2 Construction and application of recombinant Pichia pastoris displaying PET decomposing enzyme on the cell surface, including the following steps:
[0111] chemically synthesized the nucleotide sequence of the improved PET decomposing enzyme shown in SEQ ID No.1;
[0112] Design upstream primer P-F and downstream primer P-R according to the sequence of SEQ ID No.1;
[0113] P-F: 5'-3'CCGCTCGAGAAAAGAGATTACAAGGATGACGACGATAAGCAAAACCAATCCTTATGCCCGTG
[0114] P-R: 5'-3'ACTACCACCTCTCTCCACTACCTCCACCACCGCTACAGTTGGCGGTACGAA
[0115] Perform PCR, and perform 1% agarose gel electrophoresis on the PCR reaction product to recover the target gene fragment to obtain the PET decomposing enzyme gene.
[0116] According to GCW51 (NCBI Reference Sequence: XP_002493782), after predicting its signal peptide, design upstream primer 51-F and downstream primer 51-R;
[0117] 51-F: 5'-3'GGTGGTGGAGGTAGTGGAGGAGGTGGTAGTGATGACGATGACTCATTAC
[0118] 51-R: 5'-3'CCGGAATTCCTAGATCAATAGGGCAATG...
Embodiment 3
[0189] Example 3 Construction and application of recombinant Pichia pastoris displaying PET decomposing enzyme on the cell surface, including the following steps:
[0190] chemically synthesized the nucleotide sequence of the improved PET decomposing enzyme shown in SEQ ID No.1;
[0191] Design upstream primer P-F and downstream primer P-R according to the sequence of SEQ ID No.1;
[0192] P-F: 5'-3'CCGCTCGAGAAAAGAGATTACAAGGATGACGACGATAAGCAAAACCAATCCTTATGCCCGTG
[0193] P-R: 5'-3'ACTACCACCTCTCTCCACTACCTCCACCACCGCTACAGTTGGCGGTACGAA
[0194] Perform PCR, and perform 1% agarose gel electrophoresis on the PCR reaction product to recover the target gene fragment to obtain the PET decomposing enzyme gene.
[0195] According to GCW61 (NCBI Reference Sequence: XP_002494322), after predicting its signal peptide, design upstream primer 61-F and downstream primer 61-R;
[0196] 61-F: 5'-3'GGTGGTGGAGGTAGTGGAGGAGGTGGTAGTAACAACCTATCAAACGAGAGT
[0197] 61-R: 5'-3'ATAGTTTAGCGGCCGCTTAAATCA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com