Method for activating expression of biosynthetic gene cluster of microbial recessive secondary metabolites
A technology for secondary metabolites and biosynthesis, applied in the fields of microbiology and genetic engineering, which can solve the problems of low universality, difficult to predict results, and low efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
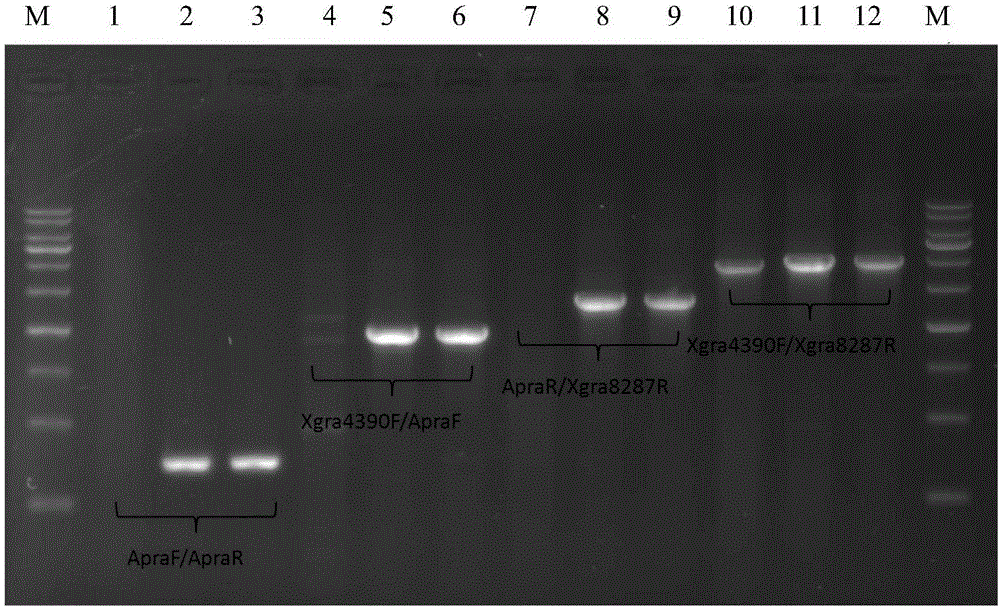
[0050] Inactivation of major secondary metabolite biosynthesis gene clusters by deletion of regulatory genes
[0051]Using Streptomyces vietnamensis GIMV4.0001 genomic DNA as a template, using primers Xgra4390F (5'-ACGCCAAGGAGGTGCTCACGAC-3', as shown in SEQ ID NO.1) / Xgra8287R (5'-CTTCCTGTCCGGGCACAA-3', as shown in shown in SEQ ID NO.2) to perform PCR to amplify a 3898bp fragment containing the two-component regulatory gene gra-orf11 and its upstream and downstream sequences. LA-Taq was used for PCR, and a 25 μL reaction solution containing 10 ng of genomic DNA fragments, 20 pmol of primers Xgra4390F and 20 pmol each of Xgra8287R was prepared according to the attached instructions, and carried out under the following conditions: pre-denaturation at 94°C for 3 minutes, denaturation at 94°C for 35 seconds, Anneal at 64°C for 1 minute, extend at 72°C for 3.5 minutes, do denaturation-annealing-extension cycle 30 times, store at 72°C for 10 minutes.
[0052] The PCR amplification p...
Embodiment 2
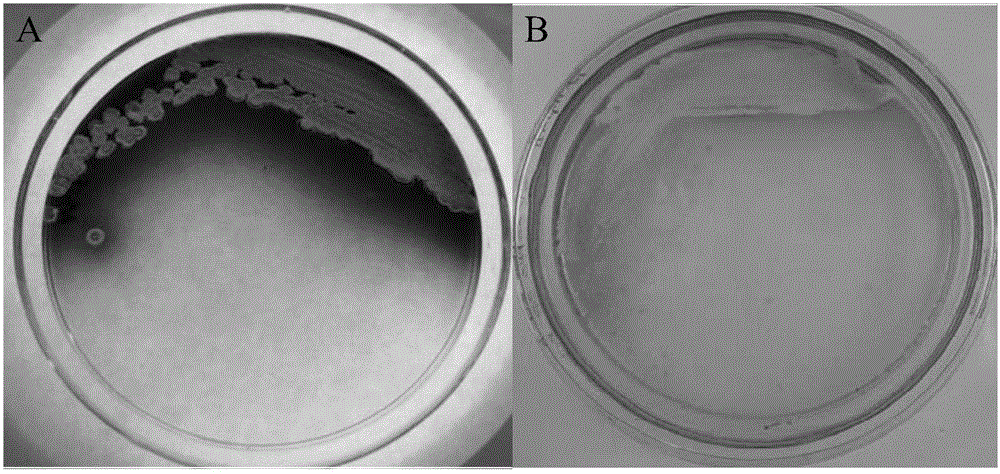
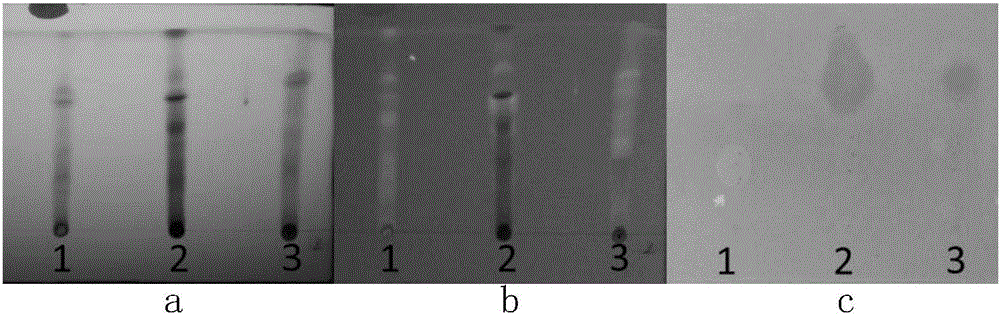
[0058] Activation of microbial mutants activates recessive secondary metabolite biosynthesis gene clusters by altered culture conditions
[0059]Group A experiment: Streptomyces vietnamensis Δgra-orf11 strain and Streptomyces vietnamensis GIMV4.0001 as a control were inoculated in Gaoshi Synthetic No. 1 Medium, Potato Glucose Medium, Sucrose Ca In the eight kinds of culture mediums including Shi's medium, calcium chloride medium, ISP3, YD, YMS, and YEME, shake culture at 30°C and 200 rpm for 7 days. Group B experiment: Another Streptomyces vietnamensis Δgra-orf11 strain and Streptomyces vietnamensis GIMV4.0001 as a control were inoculated in the above-mentioned eight kinds of media respectively, at 30°C, 200 rpm , after 3 days of shaking culture, 42°C, 200 rpm, shaking culture for 3 hours, then transfer to 30°C, 200 rpm, and continue shaking culture until the 7th day. Group C experiment: take the above eight kinds of culture medium, add 5% dimethyl sulfoxide (DMSO), ethanol a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com