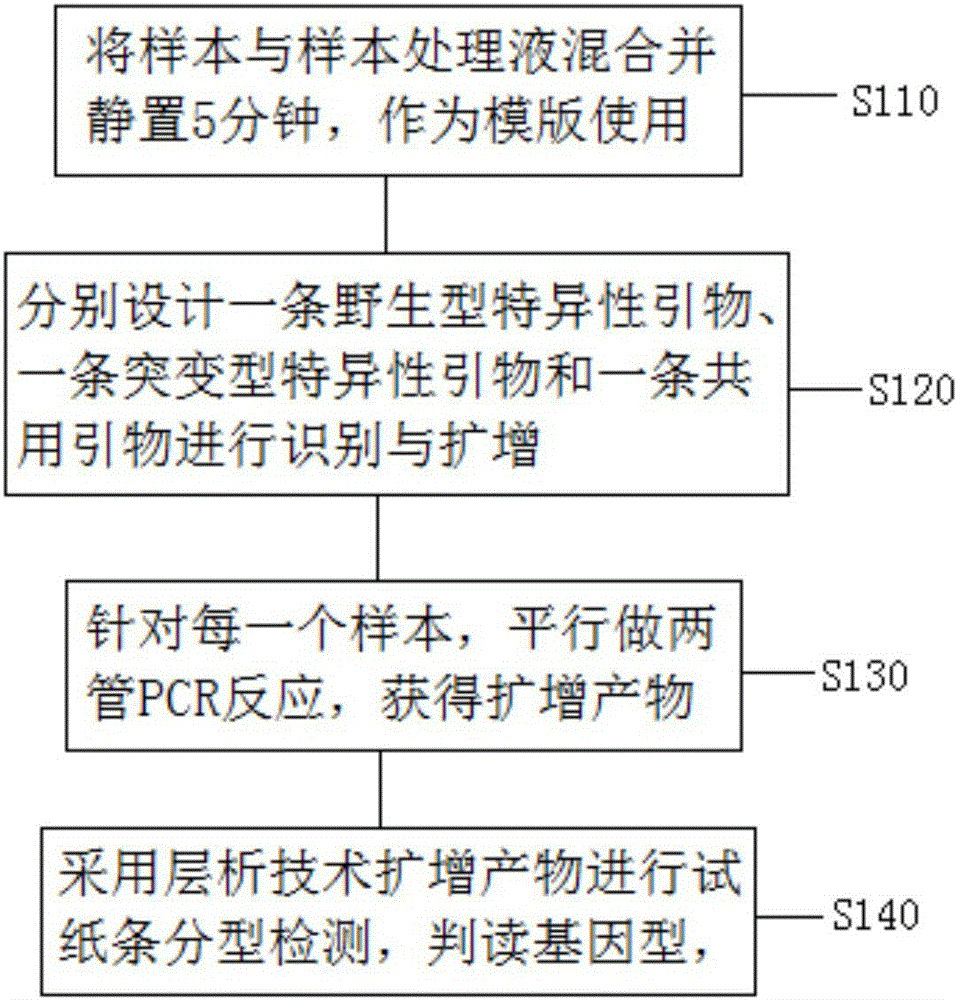
Extraction-free direct-amplification rapid detection method for SNP typing of human methylenetetrahydrofolate reductase (MTHFR) gene via test strips
A detection method and technology of test strips, applied in the field of genetic detection, can solve the problems of being unsuitable for promotion and use in clinical hospitals, increasing sample cross-contamination, and wasting a lot of time, and achieving the effects of being easy to promote and use, reducing cross-contamination, and saving costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] 1. Take a small amount of whole blood sample and add it to a PCR centrifuge tube, use a pipette to absorb 10 μL of the sample treatment solution, add it to the centrifuge tube, and let it stand for 5 minutes for later use;
[0048] 2. Take the two PCR centrifuge tubes A and B and add the following substances respectively:
[0049]
[0050] 3. Put the two tubes A and B into the PCR instrument, and proceed according to the following amplification procedure:
[0051] 31 cycles of UNG enzyme action at 50°C for 2min, pre-denaturation at 95°C for 3min, denaturation at 94°C for 5s, annealing at 60°C for 10s, extension at 72°C for 30s, 31 cycles, and final extension at 65°C for 10min. Total time: 1h20min.
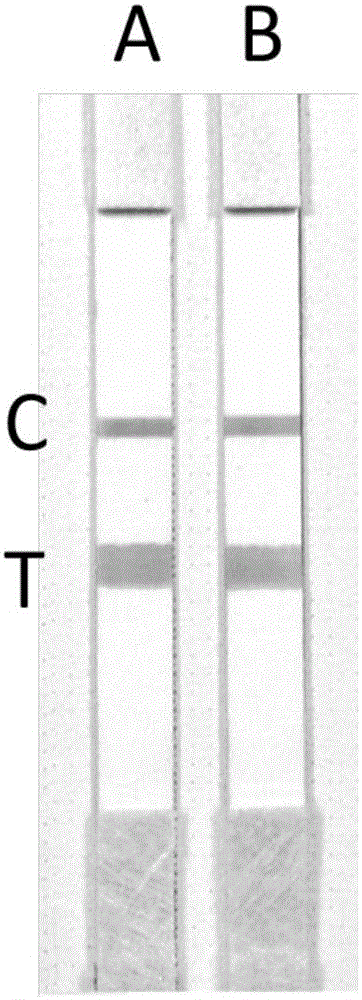
[0052] 4. Drop the amplification products of tubes A and B onto two test strips respectively, the results are as follows: figure 2 shown. according to figure 2 According to the color interpretation of the middle T line, the sample is a heterozygous mutant of MTHFR C...
Embodiment 2
[0054] 1. Take a small amount of oral swab sample and add it to the PCR centrifuge tube, use a pipette to absorb 10 μL of the sample treatment solution, add it to the centrifuge tube, and let it stand for 5 minutes for later use;
[0055] 2. Take C and D two PCR centrifuge tubes and add the following substances respectively:
[0056]
[0057] 3. Put the two tubes C and D into the PCR instrument, and proceed according to the following amplification procedure:
[0058] 31 cycles of UNG enzyme action at 50°C for 2min, pre-denaturation at 95°C for 3min, denaturation at 94°C for 5s, annealing at 60°C for 10s, extension at 72°C for 30s, 31 cycles, and final extension at 65°C for 10min. Total time: 1h20min.
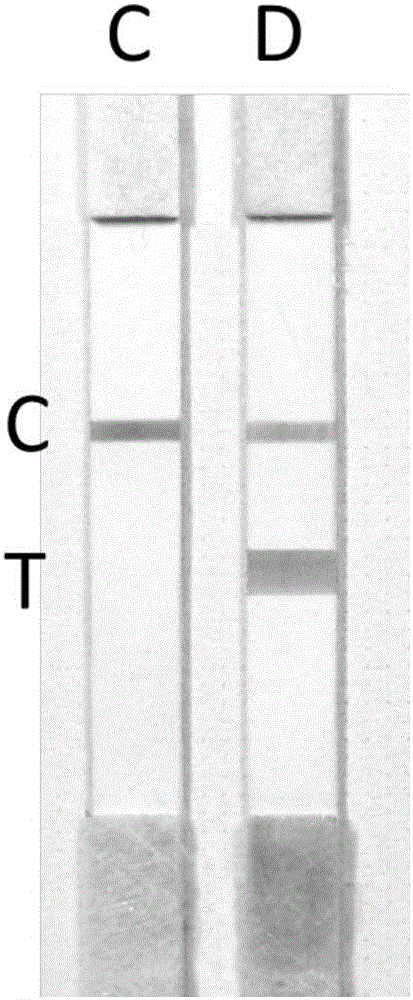
[0059] 4. Drop the amplified products of tubes C and D onto two test strips respectively, and the results are as follows: image 3 shown. according to image 3 The middle T line color interpretation, the sample is MTHFR C677T wild type.
Embodiment 3
[0061] 1. Take a small amount of fingertip blood spot sample and add it to a PCR centrifuge tube, use a pipette to absorb 10 μL of the sample treatment solution, add it to the centrifuge tube, and let it stand for 5 minutes for later use;
[0062] 2. Take the two PCR centrifuge tubes A and B and add the following substances respectively:
[0063]
[0064] 3. Put the two tubes A and B into the PCR instrument, and proceed according to the following amplification procedure:
[0065] 31 cycles of UNG enzyme action at 50°C for 2min, pre-denaturation at 95°C for 3min, denaturation at 94°C for 5s, annealing at 60°C for 10s, extension at 72°C for 30s, 31 cycles, and final extension at 65°C for 10min. Total time: 1h20min.
[0066] 4. Drop the amplification products of tubes A and B onto two test strips respectively, the results are as follows: Figure 4 shown. according to Figure 4 According to the color interpretation of the middle T line, the sample is a homozygous mutant of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com