Salmonella flagellin fusion protein as well as encoding gene and application thereof
A Salmonella and fusion protein technology, applied in the field of genetic engineering, can solve the problems of mismatched vaccine strains and easy mutation of antigens
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The selection and optimization of embodiment 1 gene
[0042] The Salmonella typhimurium flagellin gene (FliC) 1521bp and the cholera enterotoxin B subunit gene (CTB) 375bp were selected, and the codons in the above two genes were transformed into potato-preferred codons through a codon preference library. The analyzed genes were translated into protein sequences using DNA star software, and protein alignment analysis was performed by pBLAST in NCBI. After the alignment is consistent, the splicing and construction of the gene is carried out, and the enhancing and stabilizing sequences are added before and after the fusion sequence, and the required enzyme cutting sites are added at both ends.
Embodiment 2
[0043] The fusion and construction of embodiment 2 gene
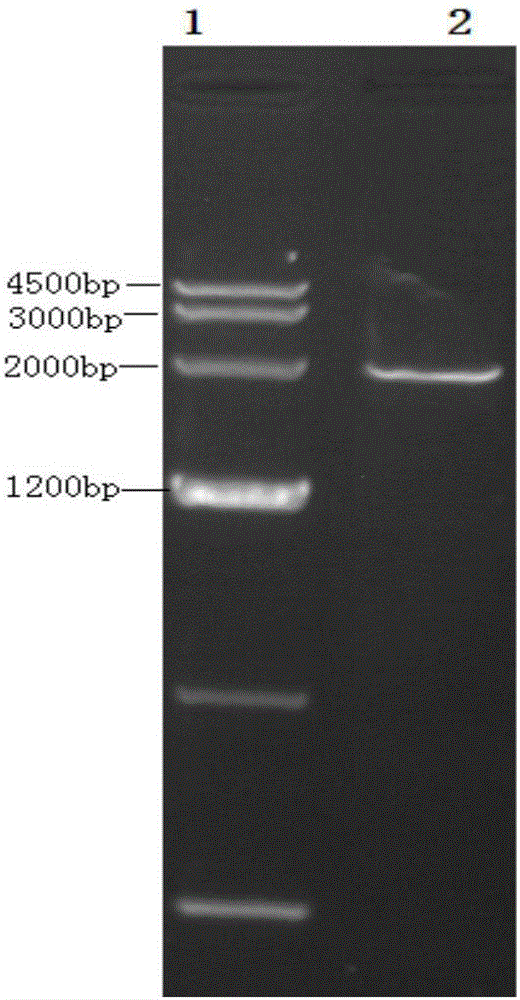
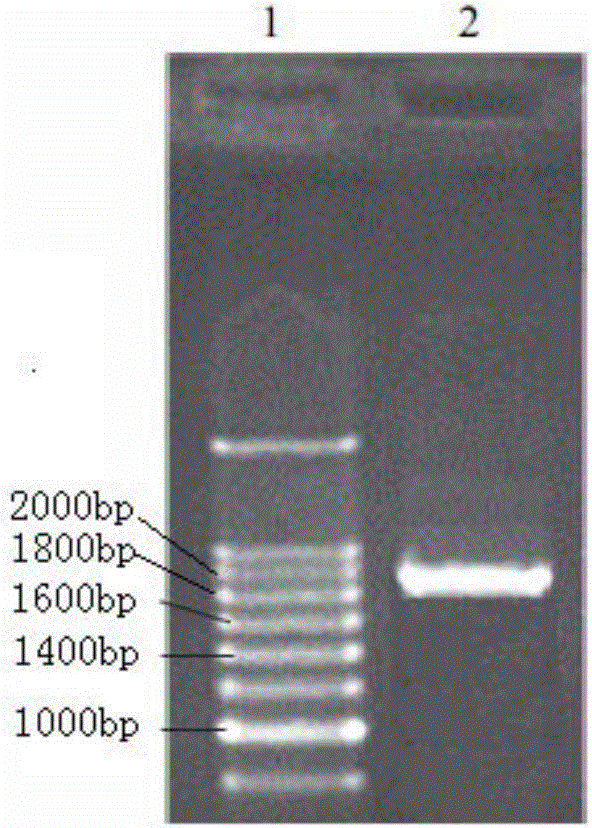
[0044] The above two genes were spliced, and the restriction sites were analyzed with Mapdraw. Finally, add the Kozak sequence before the fused FliC and CTB gene sequences, then add the endoplasmic reticulum retention peptide, and add the required enzyme cutting sites at both ends, upstream Ndel, Sal1, and downstream Sac1. The optimized gene was sent to the company for synthesis, and the full length of the synthetic gene was 1932bp. The synthetic sequence was linked into T vector pMD19(Simple), and finally T-Vector pMD19(simple)-fliC-CTB was synthesized. The base sequence of the fusion gene is shown in SEQ ID NO.2, its ORF is 1908bp long, and the base sequence is shown in SEQ ID NO.3. The gene encodes 635 amino acids, including a stop codon.
Embodiment 3
[0045] Embodiment 3 plant expression vector construction
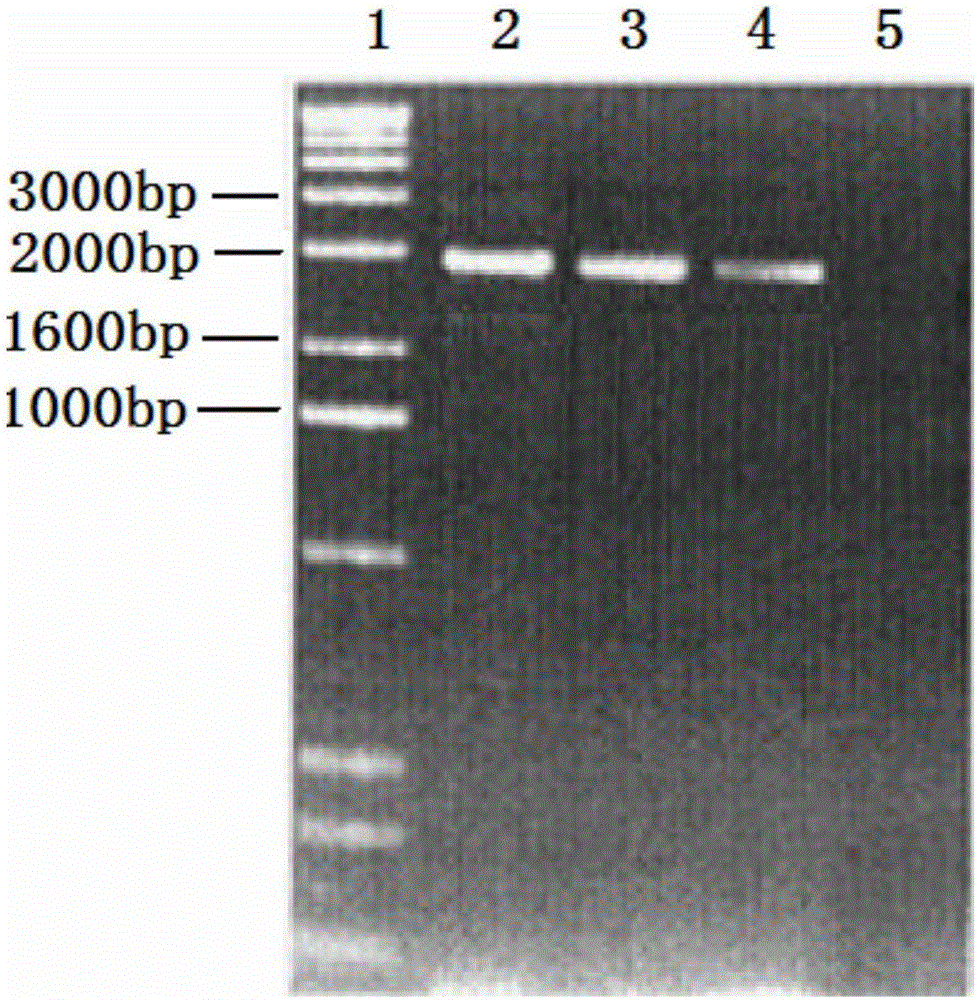
[0046] T-Vector pMD19(simple)-fliC-CTB recombinant plasmid and pRI 101-AN DNA empty vector were respectively digested with Ndel+Sac1 double enzymes, 1% agarose electrophoresis, the voltage was 3-5v / cm, the target fragment FliC- After CTB was separated from the pRI 101-ANDNA vector, it was separated and cut with a surgical blade under ultraviolet light, and the target fragment and vector were recovered with an agarose recovery kit. The target fragment and the carrier were ligated with T4 ligase at an equimolar ratio of 1:1. The ligation reaction was carried out at 16°C for 1 hour, and the reaction was terminated at 70°C. Transform the ligation product into E. coli DH5α, spread 200 μL of the transformed bacteria solution on the flat plate containing kanamycin LB, spread it evenly with a spreader, and culture it horizontally in a 37°C incubator for 1 hour, then turn the plate upside down Incubate for 12 hours. The clon...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com