Molecular biological method for rapidly identifying Hoolock leuconedys and Nomascus leucogenys
A molecular biology and gibbon technology, applied in the field of molecular biology, can solve the problems of identifying difficult criminals with materials, distinguishing and identifying Eastern White-browed Gibbons and Northern White-cheeked Gibbons, etc., to protect biodiversity, avoid transportation difficulties, and reduce transportation. difficult effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
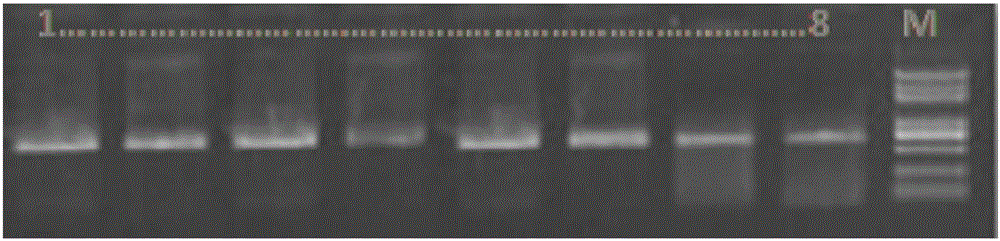
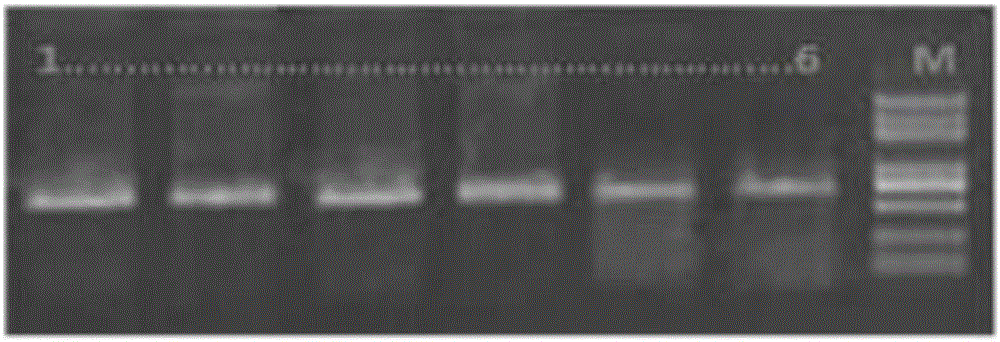
Image
Examples
Embodiment 1
[0042] 1. Sample collection and storage
[0043] Four samples were used in the experiment of the present invention, including two eastern hoomo gibbons, "Bei Bei" and "Gua Gua", and two northern white-cheeked gibbons, "Le Le" and "Wei Wei". See Table 1 for sample details.
[0044] Table 1. Conditions of the samples used in the experiment of the present invention.
[0045]
[0046]
[0047] 1.1 Blood samples
[0048] The samples were collected from fresh venous blood from the eastern hoolock gibbon "Bei Bei" and the northern white-cheeked gibbon "Lele". Eastern Hoolock Gibbon "Bei Bei", female, 9 years old, was rescued by the Yunnan Provincial Wildlife Shelter and Rescue Center in Dehong Prefecture, Yunnan Province in 2015. The northern white-cheeked gibbon "Lele", female, 18 years old, was rescued by the Yunnan Provincial Wildlife Shelter and Rescue Center in Xishuangbanna Prefecture, Yunnan Province in 2006. The morphological identification of the two gibbons was co...
Embodiment 2
[0115] The object to be tested is a piece of muscle tissue, with an area of about 0.3×0.4cm and a weight of about 1.6g. It was provided by the Forest Public Security Bureau of Yunnan Province. It is suspected to be a species of Eastern Hoolocks Gibbon and Northern White-cheeked Gibbon.
[0116] Take about 100 mg of the object to be tested in the sample tube, and cut the tissue pieces into pieces with clean scissors. Directly use the DNA extraction kit (Shanghai Huashun Biological Engineering Co., Ltd.) to extract. During the extraction process, a little more RNase A needs to be added to minimize RNA residue to ensure DNA quality.
[0117] The PCR amplification reaction system, PCR reaction conditions and gene sequence determination were all consistent with the samples in Example 1.
[0118] Compare the COI gene sequence of the test object directly with Table 5. For example, first look at the 10th nucleotide position, and the result shows that the base to be tested is "A"; ...
Embodiment 3
[0120] The object to be tested is a piece of fur, with an area of about 1.3×1.6cm, provided by the Forest Public Security Bureau of Yunnan Province. It is suspected to be a species of eastern hooze gibbon and northern white-cheeked gibbon.
[0121] Hair DNA extraction, PCR amplification reaction system, PCR reaction conditions and gene sequence determination are all consistent with the samples in Example 1.
[0122] Compare the COI gene sequence of the test object directly with Table 5. For example, first look at the 10th nucleotide position, and the result shows that the base to be tested is "G"; secondly, look at the 42nd nucleotide position, and the base to be tested is "C". By analogy, we can finally conclude that the source of the fur tissue sample is the northern white-cheeked gibbon.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com