A detection kit for multiplexed amplification of 21 short tandem repeats using a double fluorescent labeling method
A technology of short tandem repeats and detection kits, applied in the field of molecular genetics, can solve the problems of difficult sensitivity and achieve the effects of high fluorescence efficiency, strong amplification specificity, and good species specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
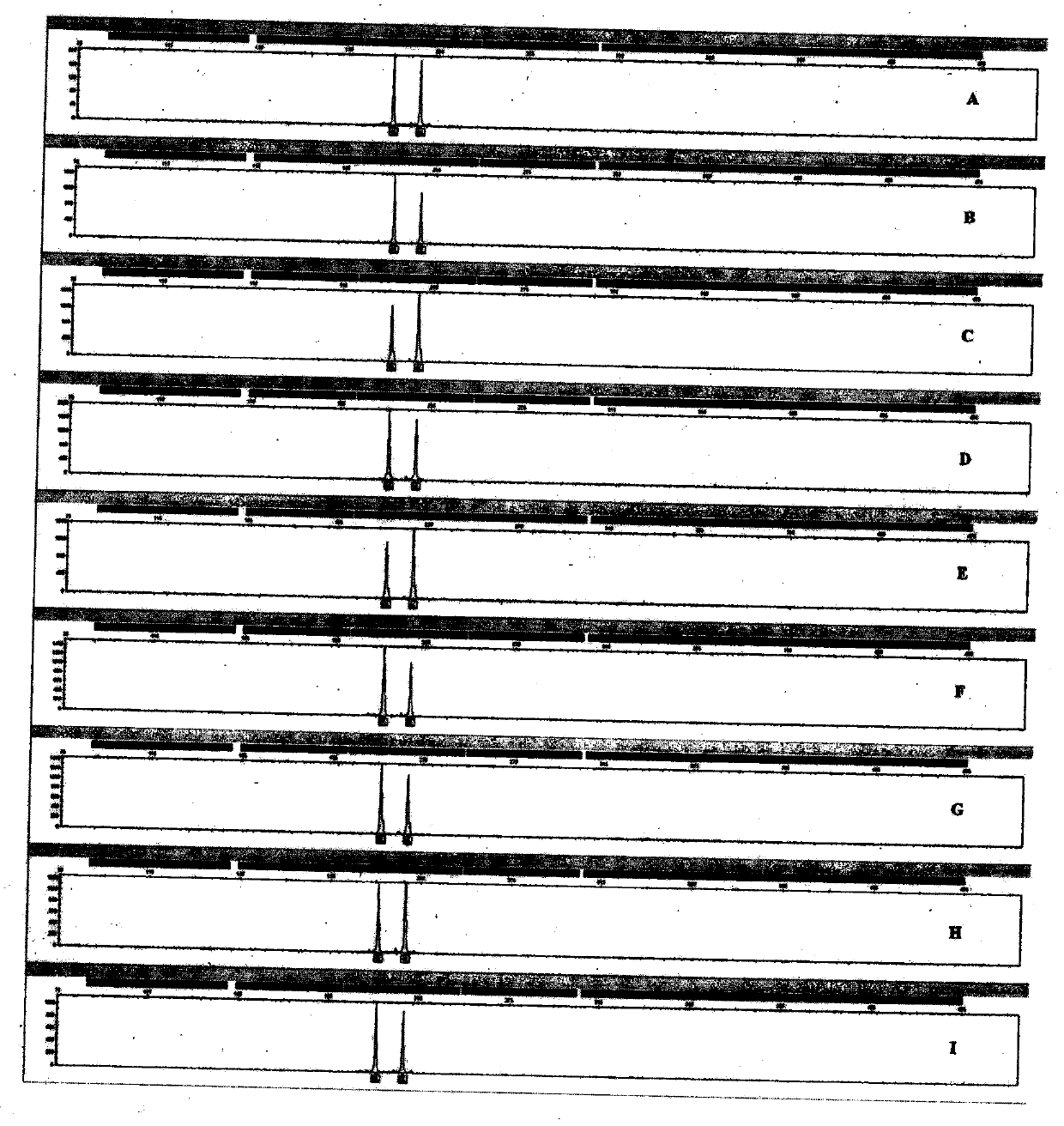
[0046] Example 1, Primer Design and Determination of the Position of the Fluorescent Label on the Primer Sequence
[0047] The system amplifies a total of 21 STR loci, including Amelogenin, D3S1358, vWA, D7S820, CSF1PO, D8S1179, D21S11, D16S539, TH01, D13S317, TPOX, D18S51, D5S818, FGA, D2S1338, D19S433, D1S1659, D12S D, D1S1659, D12S Penta E and D6S1043.
[0048] Specific primers were designed for the above 21 loci on the flanks of their repeated sequences. Primers were designed using Oligo7 software, and the Tm value of each primer was close to 60°C. The maximum range of the length of the amplified product is 75-500bp. Each pair of primers is tested and optimized until the amplified peak with sharp peak shape and high peak height is obtained. After the multiplex amplification test, between 56°C and 63°C, there is no non-specific amplification, all have target peaks, and no other interactions or cross-reactions occur. No amplification of DNA from dogs, pigs, cattle, sheep,...
Embodiment 3
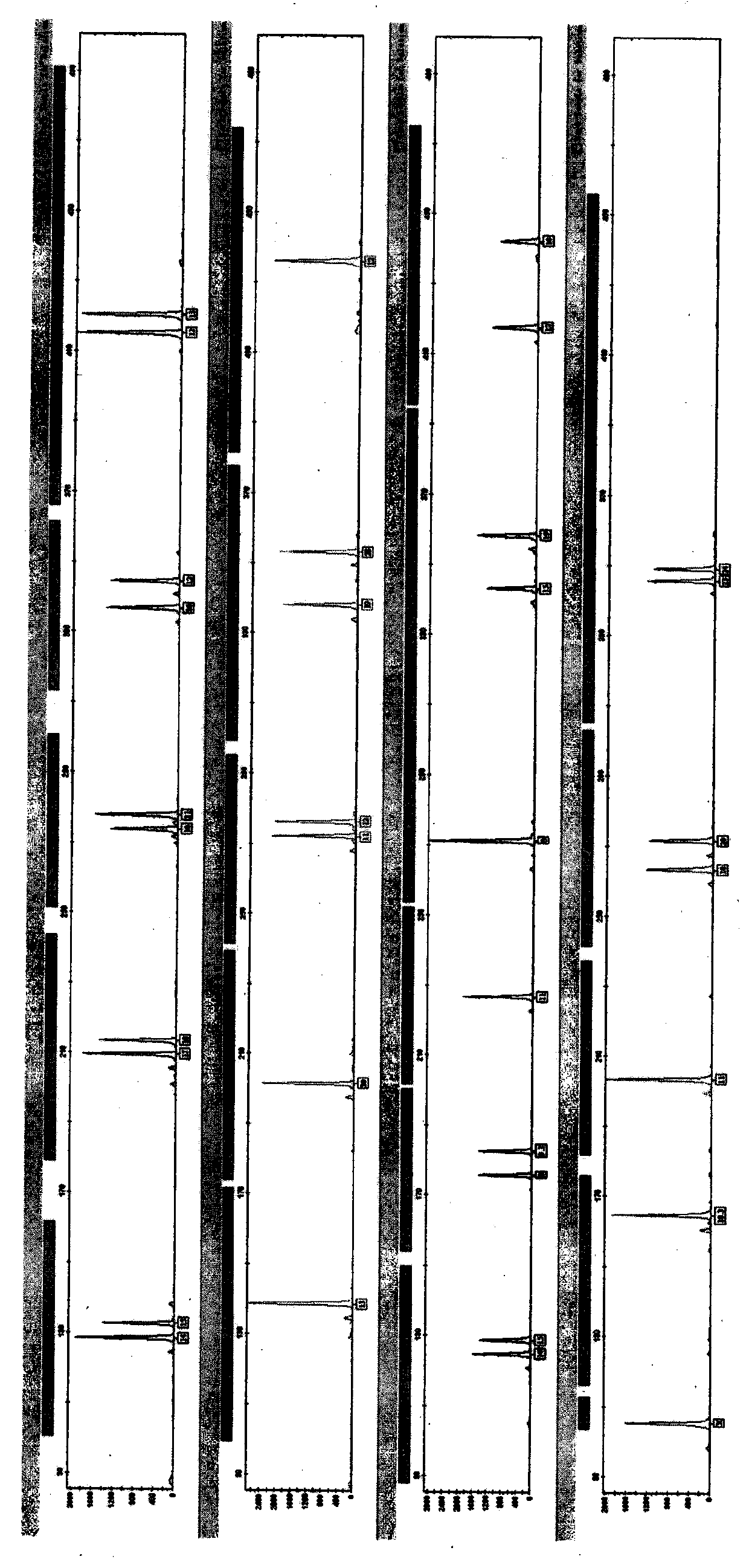
[0070] Embodiment 3, compare the consistency of typing of the present invention and other commercial kits
[0071] In order to detect the consistency of typing between the present invention and other commercial kits, this embodiment selects Microreader TM 21 Direct ID System was used as a reference object, and 1316 samples (including 663 irrelevant individuals) were amplified using the present invention and 21 Direct ID kit respectively, and the products were typed by ABI 3100 genetic analyzer capillary electrophoresis, and the type The results were analyzed and compared. 21Direct ID kit is operated according to its instructions, and the steps of the present invention are as follows:
[0072] 3.1. Polymerase chain reaction (PCR) amplification
[0073] 1) Take the buffer, primer mixture, and Taq enzyme, and prepare a mixture according to the table below, shake and mix well, and then dispense into PCR reaction tubes, 25 μL per tube.
[0074] this invention:
[0075]
[00...
Embodiment 4
[0089] Embodiment 4, the sensitivity comparison of the present invention and conventional labeling method amplification system
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com