Stenotrophomonas maltophilia outer membrane protein and application thereof
A protein and sequence technology, applied in antibacterial immunoglobulin, application, immunoglobulin, etc., can solve the problem of less antigen research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
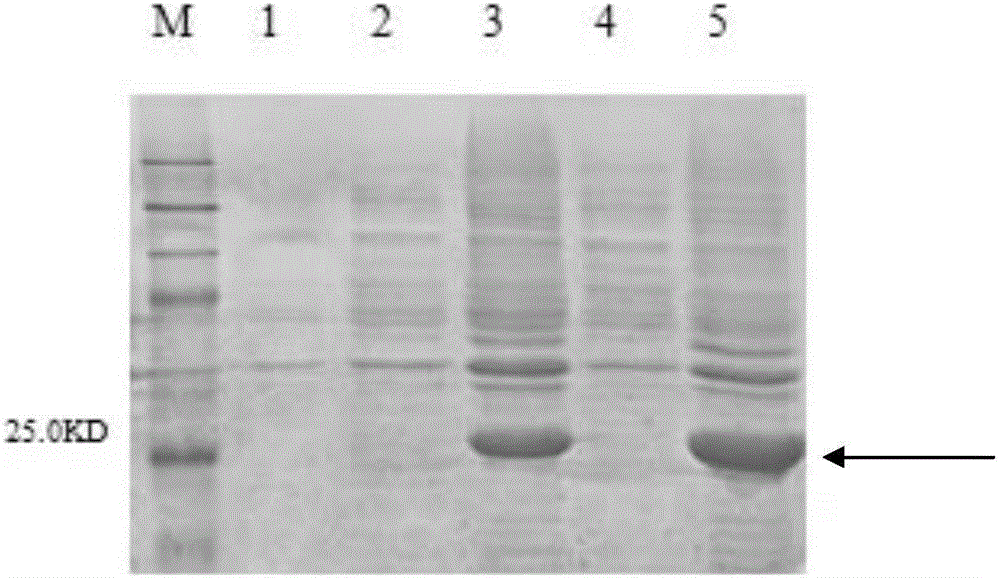
[0069] Example 1, Expression and Purification of Stenotrophomonas maltophilia Outer Membrane Protein
[0070] 1. Genomic DNA of Stenotrophomonas maltophilia K279a was extracted.
[0071] 2. Using the genomic DNA obtained in step 1 as a template, PCR amplification is performed using primers P1 and P2 to obtain PCR amplification products.
[0072] P1: 5′CG GAATTC CAGGACAGCAGTCCACCGAT-3′;
[0073] P2: 5′-CC CTCGAG TCAGAAGCGGGCACCGAT-3'.
[0074] In P1 and P2, the underline marks the restriction sites of EcoR I and Xho I, respectively.
[0075] 3. Use restriction endonucleases EcoR I and Xho I to double digest the PCR amplified product of step 2, and reclaim the digested product
[0076] 4. Digest the pET-30a(+) vector with restriction endonucleases EcoR I and Xho I to recover a vector backbone of about 5388 bp.
[0077] 5. Ligate the digested product of step 3 with the vector backbone of step 4 to obtain the recombinant vector pET-30a-Smlt4123. According to the sequencin...
Embodiment 2
[0092] Embodiment 2, preparation of antiserum by recombinant protein ompW immunization mice
[0093] Experimental mice: male BALB / c mice (6-8 weeks).
[0094] 1. The recombinant protein ompW solution obtained in step 9 and the control protein solution obtained in step 10 were quantified and diluted to 100 μg / ml respectively to obtain recombinant protein ompW dilution and control protein dilution.
[0095] 2. The experimental mice were randomly divided into two groups (10 mice in each group).
[0096] On day 0, the groups are grouped as follows:
[0097] Experimental group: Take the pre-immune serum (take blood from the eye socket, let it stand at room temperature for 60 minutes, then centrifuge at 2500rpm for 5 minutes, take the upper layer of serum, which is the pre-immune serum), and then use the recombinant protein ompW antigen emulsion I to immunize the experimental mice (subcutaneously in the abdomen and back). Multi-point injection, 100μL / only).
[0098] Control group...
Embodiment 3
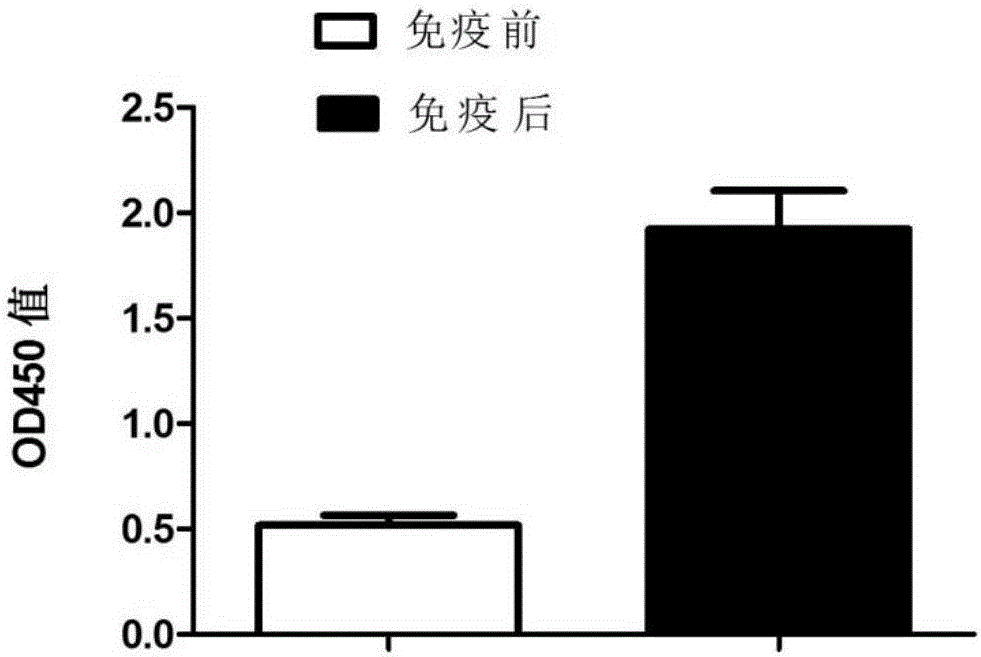
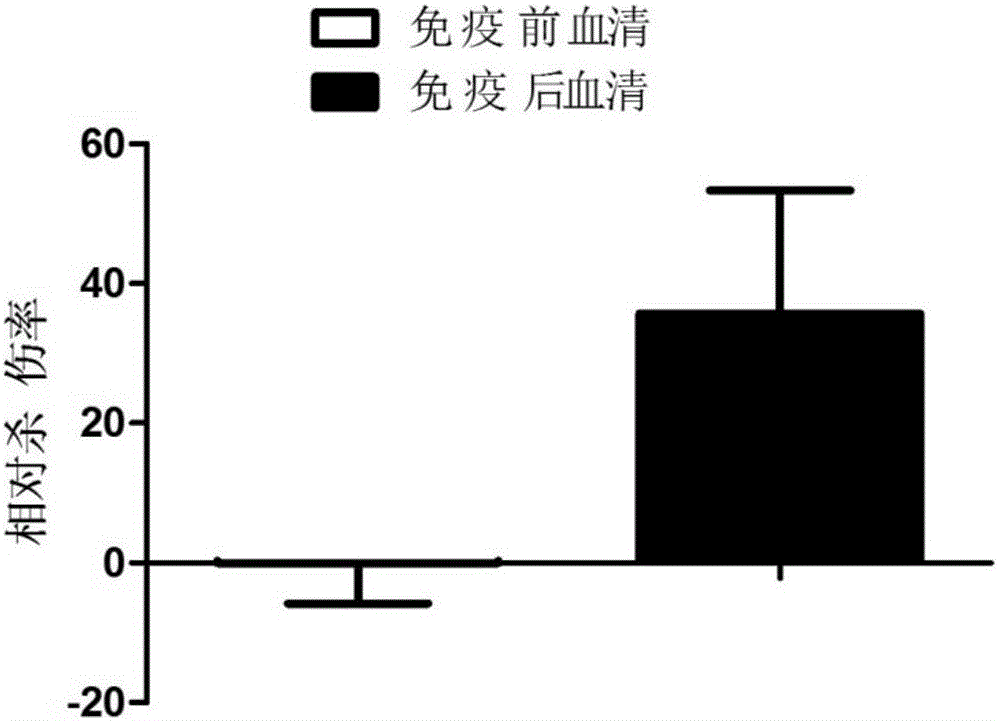
[0107] Embodiment 3, antiserum potency determination
[0108] Serum to be tested: the pre-immune serum obtained in Example 2, the post-immunization serum of the experimental group, and the post-immunization serum of the control group.
[0109] 1. Use PBST buffer to test serum 1:1×10 5 Dilute to obtain the serum dilution to be tested.
[0110] 2. The recombinant protein ompW solution obtained in Example 1 was quantified and diluted to 10 μg / ml, then added to a 96-well plate (100 μl per well), and coated at 4° C. overnight.
[0111] 3. After completing step 2, discard the solution in the well, and wash the plate with 0.5% (volume percentage) PBST solution.
[0112] 4. After completing step 3, add 100 μL of the serum dilution to be tested in step 1 to each well, and incubate at 37°C for 30 minutes.
[0113] 5. After completing step 4, discard the solution in the well, and wash the plate with 0.5% (volume percentage) PBST solution.
[0114] 6. After completing step 5, add 100 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com