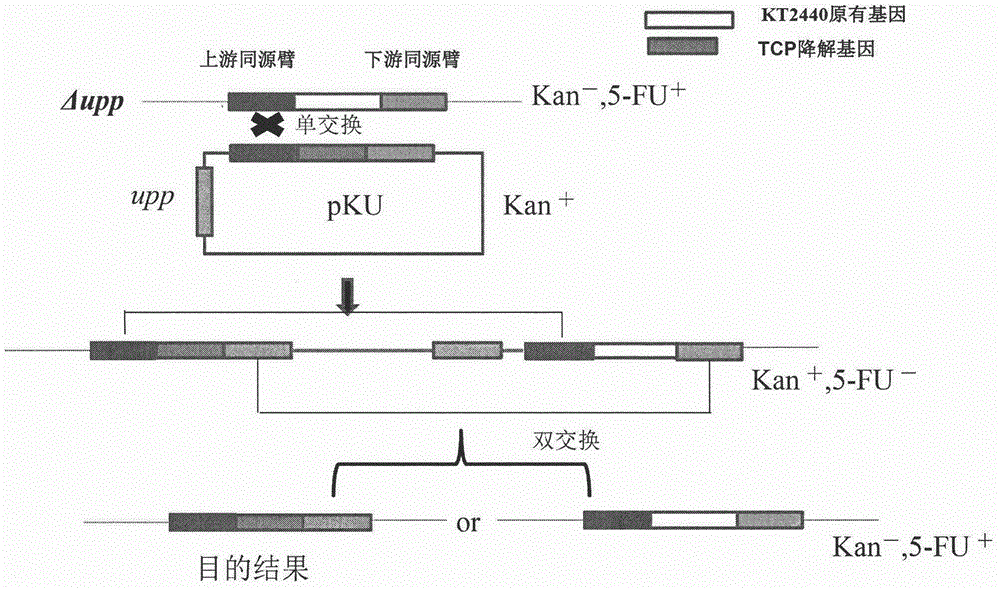
Method for constructing trichloropropane degrading bacterium by taking pseudomonas putida as original strain
A technology of Pseudomonas putida, trichloropropane, applied in the field of environmental technology restoration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment 1, take Pseudomonas putida as the construction of the trichloropropane degrading bacteria of bacterial strain
[0019] Liquid medium used in the experiment:
[0020] Luria-Bertani (LB) medium: tryptone 10g, yeast extract 5g, sodium chloride 10g, water 1L, pH7.0, sterilized at 121°C for 20 minutes for later use.
[0021] Kanamycin (Kan): Weigh 1g of Kanamycin into a 100mL beaker, add 90mL of sterilized ddH 2 O water, stir well to dissolve, dilute to 100mL, filter and sterilize, aliquot (0.5-1mL / part), store at -20°C, and set aside.
[0022] Pentafluorouracil (5-FU): Weigh 0.0375 g of pentafluorouracil into a 1.5 mL EP tube, and dissolve it in 500 μL of dimethyl sulfoxide solution at a concentration of 106 μg / mL.
[0023] The three genes dhaA31, hheC and echA in the trichloropropane degradation pathway were cloned from the host bacteria Rhodococcus rhodochrous NCIMB 13064 (GenBank accession no.AF060871.1) and Agrobacterium radiobacter AD1 (GenBank accession n...
Embodiment 2
[0027] Embodiment 2, optimize the expression of trichloropropane degradation pathway in host bacteria
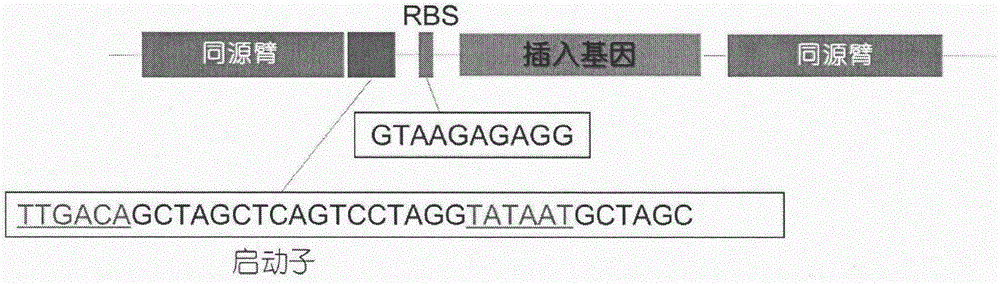
[0028] In view of the fact that none of the three genes inserted in Example 1 are transcribed, we optimized the codons of the three genes dhaA31, hheC and echA according to the codon preference of Pseudomonas putida. The optimized codon sequences are shown in SEQID NO.1, respectively. Shown in SEQ ID NO.2 and SEQ ID NO.3, the promoter and RBS in the original strain were replaced at the same time, and the promoter and RBS ( image 3 ). The strain construction and RT-PCR detection were the same as in Example 1. RT-PCR test results such as Figure 4 As shown, the three exogenous genes were all correctly transcribed.
Embodiment 3
[0029] Embodiment 3, verify the function of trichloropropane degrading engineering bacteria
[0030] Liquid media used in the experiments:
[0031] SMM medium: Na 2 HPO 4 12H 2 O 5.4g, KH 2 PO 4 1.4g, (NH) 2 SO 4 0.5g, MgSO 4 ·7H 2 O 0.2g, 2mL trace elements, 1L water, pH7.0, sterilized at 121°C for 20 minutes for later use.
[0032] Trace element solution composition: 0.1M HCl 1000mL, MnSO 4 4H 2 O 0.86g, ZnSO 4 ·7H 2 O 0.2g, CoSO 4 ·7H2 O0.28g, CuSO 4 ·5H 2 O 0.25g, FeSO 4 ·7H 2 O 3.6g, sterilized by filtration after preparation, and sterilized at 121°C for 20 minutes for later use.
[0033] The correct transcribed recombinant bacterium that obtains in embodiment 2 is with OD 600 The inoculum size of =0.05 was inserted into 100 mL of SMM medium, and was connected with 2 mM / L trichloropropane. The trichloropropane of 2mM can degrade more than 99% in 24h, proves the efficient expression ( Figure 5 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com