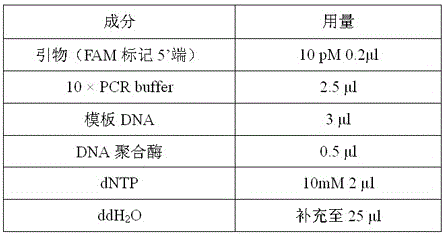
Molecular marker for identifying lyophyllum decastes, and primer and probe
A technology of lotus leaves and molecular markers, applied in the field of biological identification, can solve the problems of unreliable identification of bacterial species and difficulties in taxonomy, etc., and achieve the effects of short detection time, accurate identification results, and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Extraction of genomic DNA from Umbrella lily.
[0034] The Agaric mushroom-like material was collected from the Guanxian Mountain deciduous broad-leaved forest belt in the Xilan area of Xinzhou, Shanxi Province, and it was identified as A. Lyophyllum decastes (Fr.) Singer).
[0035] Using the tissue separation method, the whole fruiting body was wiped and disinfected with 75% ethanol, then a small piece of tissue was cut from the cap, stipe, gill and root with a scalpel, and the cut tissue was soaked in 75% ethanol for 30s. Rinse with sterile water, inoculate in PDA medium (200 g of potato, 20 g of glucose, 15 g of agar, 1 L of water) sterilized at 121° C. for 30 min. The inoculated medium was placed in a constant temperature incubator at 27°C, and the mycelial growth was checked every 24 hours.
[0036] After 8 days of culture, the hyphae were collected and the total fungal DNA was extracted with the SIGMA Fungal Genome Extraction Kit. The extraction m...
Embodiment 2
[0037] Example 2: Determination of ITS-specific molecular markers.
[0038] Using the total DNA obtained in Example 1 as a template, the DNA was amplified by PCR using the universal primers ITS1 / ITS4, and the PCR product was subjected to capillary sequencing to obtain detailed sequence information.
[0039] The sequencing results are compared and analyzed in the whole nucleic acid database in NCBI, and the fragments with high conservation are obtained by screening, and the results of the selected different sequences are compared and selected, and finally a characteristic sequence in the sequencing results is determined. The amino acid sequence of the specific gene fragment of Umbellifera is shown in SEQ ID NO.1.
[0040] After the homology comparison search, it is determined that the selected target sequence is a DNA sequence with high specificity, which can be used as the target sequence detected by the gene chip.
Embodiment 3
[0041] Example 3: Design of primers and nucleic acid probes.
[0042] According to the target sequence determined in Example 2 as a molecular marker, according to the design principles of primers and nucleic acid probes, the special primers shown in SEQ ID NO.2 and SEQ ID NO.3, and the nucleic acid probe shown in SEQ ID NO.4 were designed. Needle.
[0043] Primer 1: 5'Hex-ATGTCTTTACATACCCCATATG-3'.
[0044] Primer 2: 5'-CAAAAGTAAAGAAGTTGTCCTTA-3'.
[0045] Nucleic acid probe: 5'NH 3 -TTTTTTTTTTTTCAACCCCCACATCCAAACCTAACCAAAC-3'.
[0046] Wherein, the 5' end of the primer 1 is labeled with a fluorescent reporter group Hex; the 5' end of the nucleic acid probe is connected with an amino group, and the nucleic acid probe is subjected to amination treatment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com