A prolyl endopeptidase mutant with improved catalytic activity and specific enzyme activity
A technology of prolyl endopeptidase and Aspergillus oryzae prolyl endopeptidase, which is applied in the field of enzyme engineering, can solve the problems of catalytic efficiency and low specific enzyme activity, and achieve the effect of improving catalytic efficiency and specific activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Selection of mutation sites
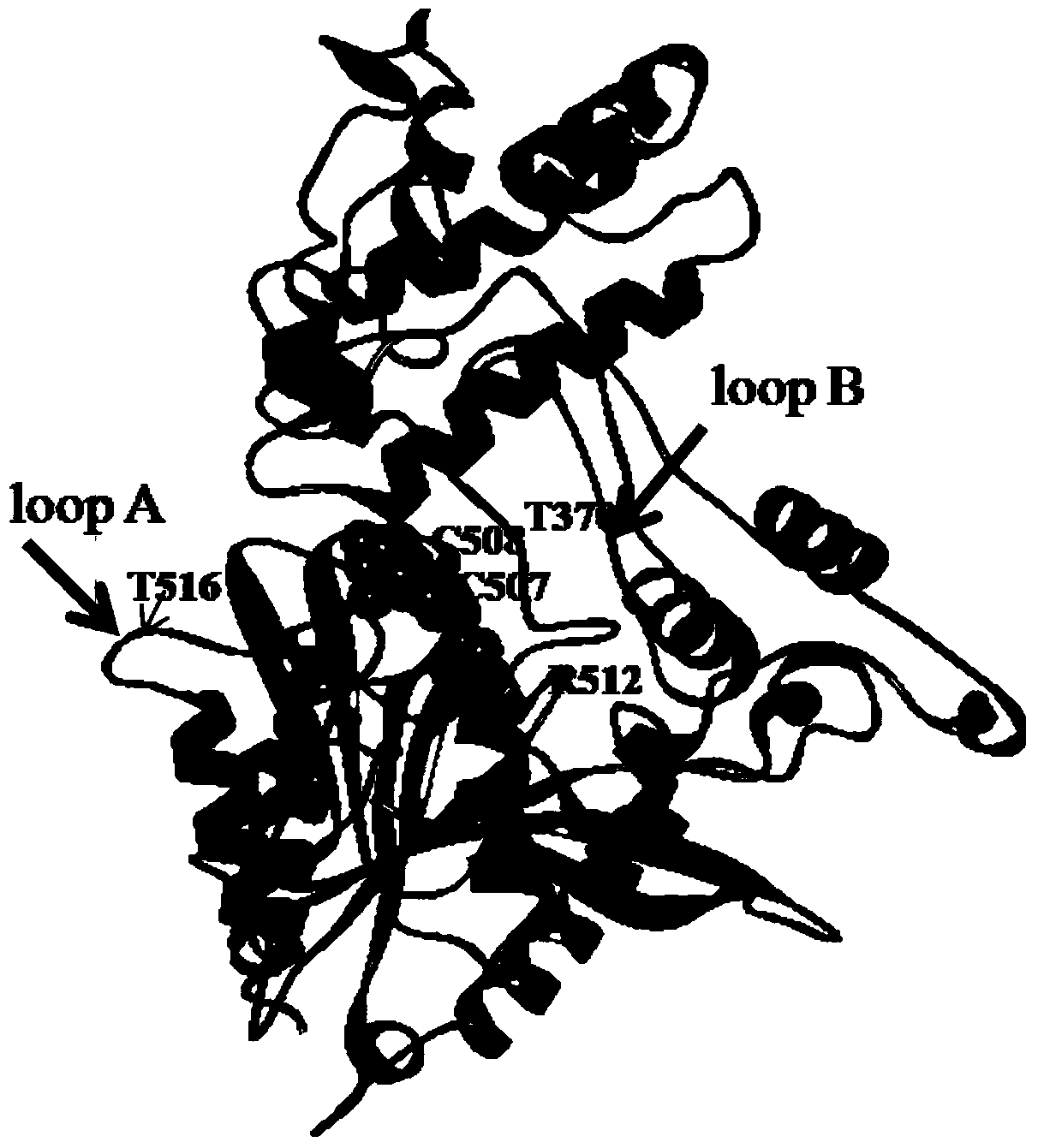
[0029] According to previous reports, the flexibility of the loop loop, an irregular area near the enzyme active center, plays a key role in the entry of the substrate into the enzyme active center and the substrate specificity, that is, the increase in the flexibility of the loop loop increases the catalytic activity of the enzyme. , combined with the three-dimensional structure obtained by homology modeling, the analysis shows that the loopA and loopB loops in the simulated structure of Aspergillus oryzae prolyl endopeptidase are both close to the surface of the active pocket ( figure 1 ), and before the substrate enters the active site, these two loops play a role in blocking the substrate from entering the active center, so these amino acids may be key amino acids. Therefore, it is possible to try to replace the hydrophilic amino acid Thr on the loop ring with other small molecular weight amino acids or hydrophobic amino acid...
Embodiment 2
[0032] Embodiment 2: the construction of the recombinant bacterium expressing mutant
[0033] Construct the expression mutant recombinant bacteria according to the following method:
[0034] (1) Construction of a recombinant plasmid with a histidine tag containing the parent Aspergillus oryzae prolyl endopeptidase: the parent Aspergillus oryzae prolyl endopeptidase gene (amino acid sequence such as SEQ ID NO.1, nucleotide The sequence is shown in SEQ ID NO.2) was cloned into pPIC9K to obtain the recombinant plasmid pPIC9K-AO-PEP, and then pPIC9K-AO-PEP was used as a template, and the F and R with 6*His tag added as primers were used for PCR, and the correct The target fragment was digested with SnaBI and NotI, and pPIC9K was ligated to obtain an expression plasmid containing 6*his tag named pPIC9K-AO-PEP-6his.
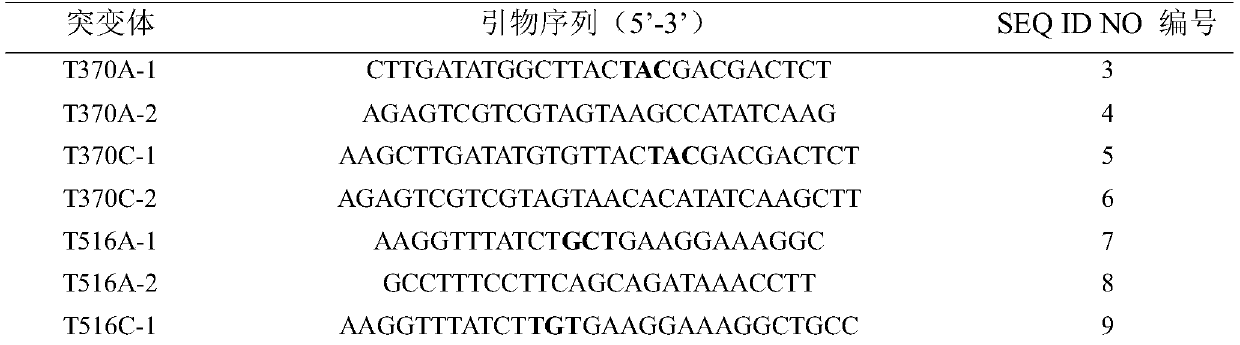
[0035] (2) Using pPIC9K-AO-PEP-6his as a template, using the primers in Table 1, using the one-step plasmid PCR-mediated site-directed mutagenesis method to construct...
Embodiment 3
[0040] Example 3: Fermentative expression and mutant purification of recombinant bacteria
[0041] Fermentative expression of recombinant bacteria and purification of mutants were carried out as follows.
[0042] (1) Fermentative expression of recombinant bacteria:
[0043] Pick a single colony of recombinant Pichia pastoris to inoculate a 250ml Erlenmeyer flask filled with 25ml BMGY medium, and culture at 30°C until the logarithmic phase OD 600 =2-6(16-18h); collect the cells by centrifugation, discard the supernatant, transfer the cells to a 500ml Erlenmeyer flask filled with 50ml of BMMY medium, place them on a shaker at 28°C and 250r / min to continue culturing, and take samples every 24h , and supplemented with 100% methanol to make the final concentration reach 1%, induced culture for 4 days, centrifuged to collect the bacteria, and carried out enzyme activity determination and SDS-PAGE electrophoresis analysis.
[0044] (2) Enzyme purification:
[0045] Take 60ml of fe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com