Carrier for in-vivo positioning mammal cell genome based on CRISPRCas9 system and application thereof
A mammalian and genomic technology, applied in vectors, nucleic acid vectors, genetic engineering, etc., can solve problems such as high difficulty, increased noise, and weak fluorescence intensity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
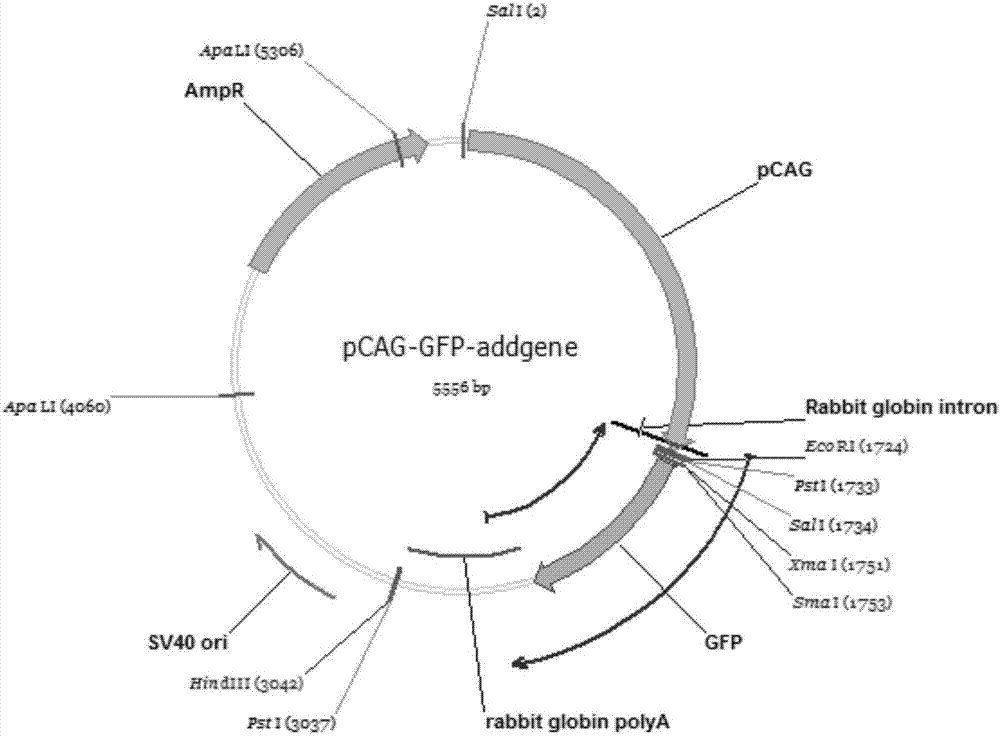
[0040] figure 1 is the pCAG-GFP-addgene plasmid map.
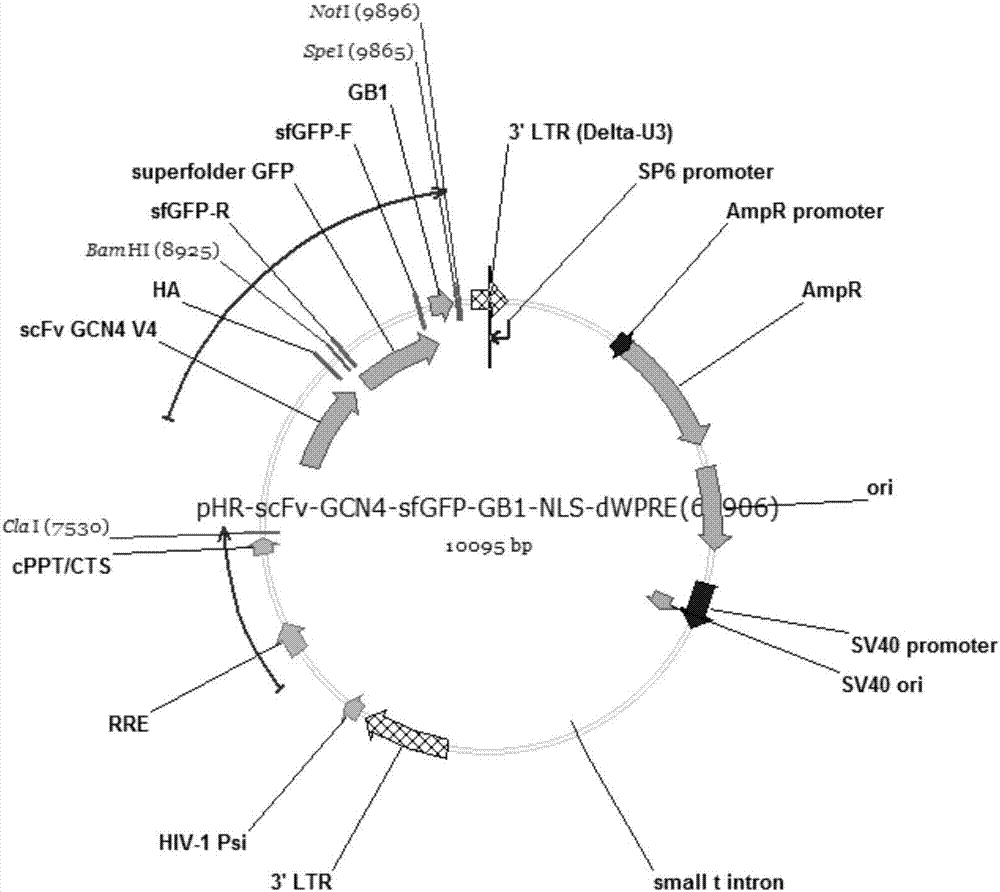
[0041] figure 2 is the plasmid map of pHR-scFv-GCN4-sfGFP-GB1-NLS-dWPRE.
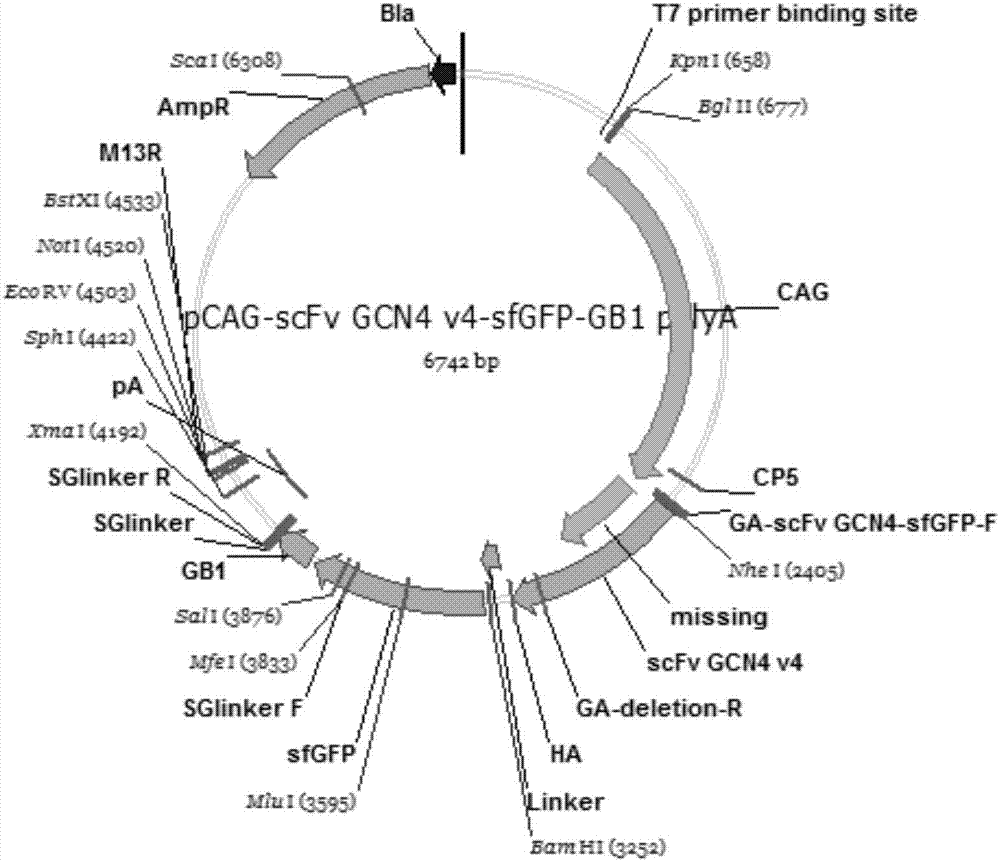
[0042] image 3 is the pCAG-scFv-GCN4_V4-sfGFP plasmid map.
[0043] Figure 4 is the pCAG-scFv-GCN4_V4-mNeonGreen plasmid map.
[0044] Figure 5 is the pCAG-scFv-GCN4_V4-3XmNeonGreen plasmid map.
[0045] Image 6 is the plasmid map of pHRdSV40-NLS-dCas9-24xGCN4_v4-NLS-P2A-BFP-dWPRE.
[0046] Figure 7is the p1U6-sgEFRNA-dCas9-24xGCN4_v4-NLS-P2A-BFP plasmid map.
[0047] Figure 8 is the p1U6-gRNA-BbsI plasmid map.
[0048] Figure 9 is the p1U6-sgEFRNA plasmid map.
[0049] Figure 10 is the p1U6-sgEFTelomere-dCas9-24xGCN4_v4-NLS-P2A-BFP plasmid map.
[0050] Figure 11 is the p1U6-sgEFTelomere plasmid map.
[0051] The specific construction steps are as follows:
[0052] The purchased pCAG-GFP (Addgene: 11150) was used as a vector, digested with EcoRI and HindIII to obtain a 4238bp fragment as a vector. Use the purchased pHR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com