Application of let-7c gene in the preparation of drugs for Alzheimer's disease
A technology for Alzheimer's disease and let-7c, applied in the field of biomedicine, to achieve the effect of inhibiting pathological changes and reducing the production of Aβ
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Karyotype analysis of DS cell lines
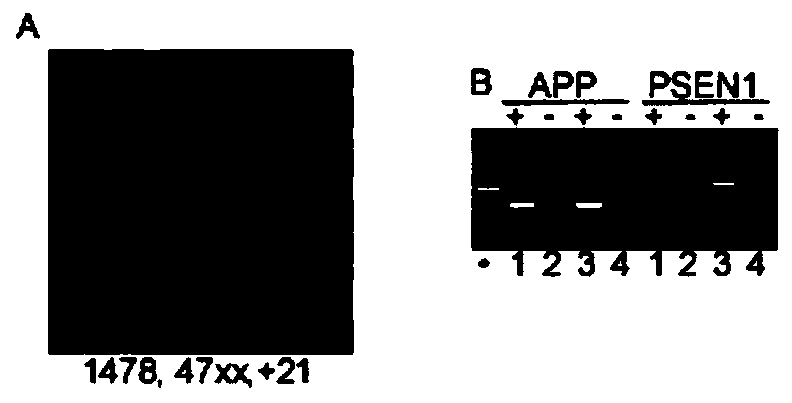
[0040] After DS cells were treated with colchicine (5ug / ml) for 3 hours, the cells were collected and treated with 0.075% KCl for 30 minutes (to expand the cells), and then fixed with fixative solution (methanol:glacial acetic acid, 3:1), and then The fixed cells were dropped onto the glass slide, treated with 0.025% trypsin for 1 minute, and finally stained with Giemsa for 10 minutes, and the karyotype analysis was carried out under the microscope. The results showed that the karyotype of the DS cell line was 47XX, +21( 1478), see figure 1 .
Embodiment 2
[0041] Example 2 Using cDNA gene chip and semi-quantitative RT-PCR to measure the expression of miRNA in DS cell line
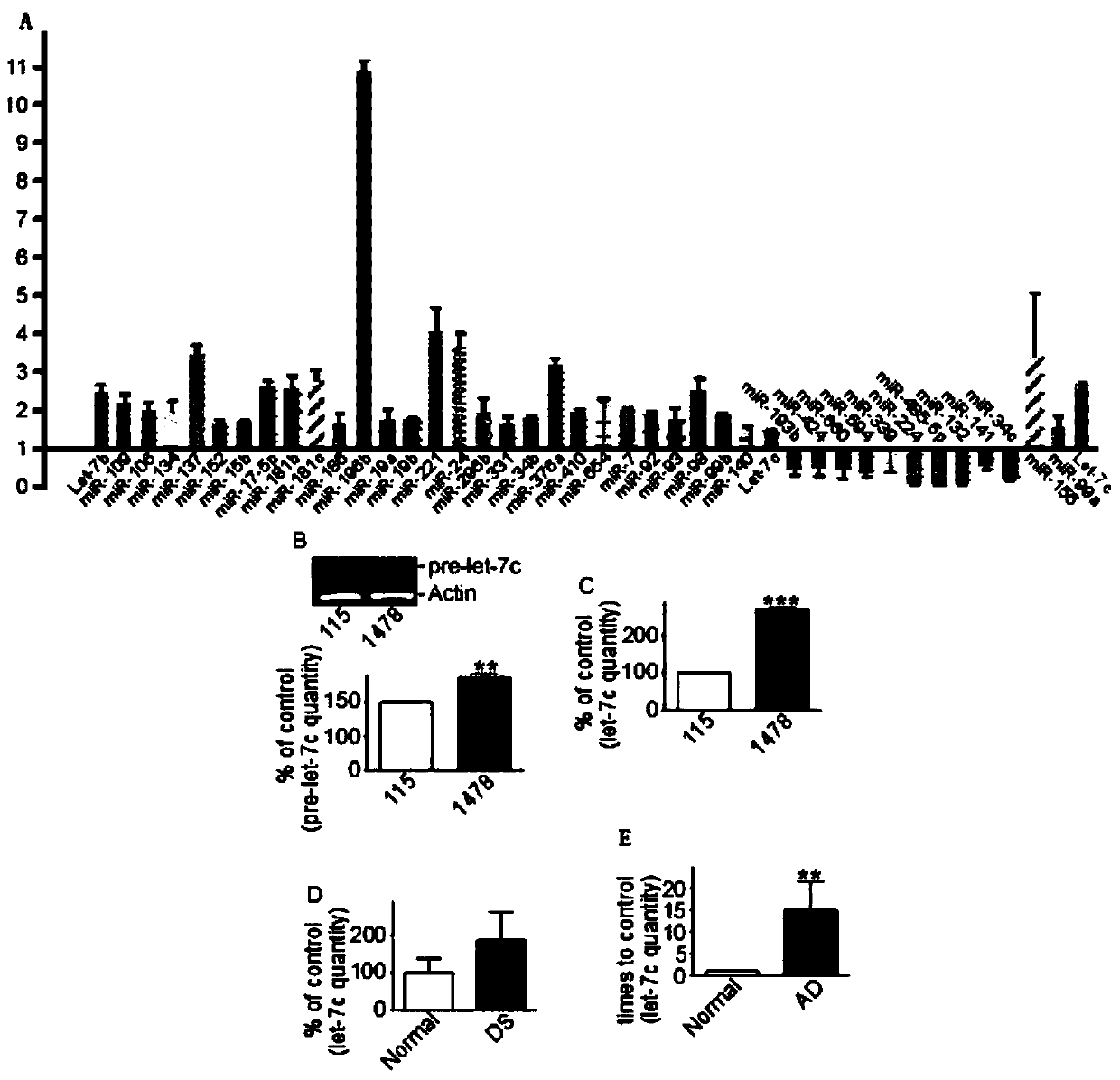
[0042] Total RNA was isolated from DS cells (UMB1478) and control cells (UMB115) using TRI-Reat reagent. According to the instructions, use Multiplex RT Human Pool4, V1for Taqman miRAN, and use the above RNA sample as a template to synthesize cDNA. Using the above cDNA as a template, the commercial TLDA Human miRNA Panel chip was used to detect the expression of different miRNAs. The results showed that the expressions of Let-7c, miR-99a, and mir-155 were up-regulated, see figure 2 a.
[0043]Using semi-quantitative RT-PCR technology, the content of pre-Let-7c (Let-7c precursor) and Let-7c in DS cells (1478) and control cells (115) were respectively determined. In addition, from DS cells and APPsw / PS1AE9 Total RNA was extracted from the brains of double mutant transgenic mice (AD mice), and the expression level of Let-7c gene was detected by semi-quantitat...
Embodiment 3
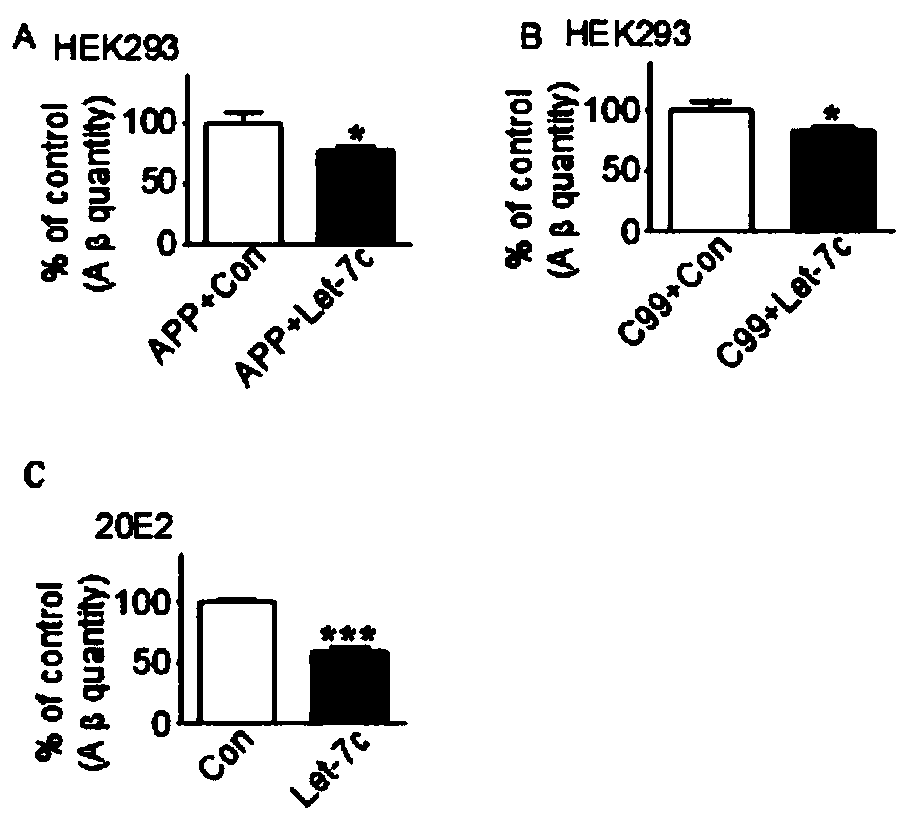
[0044] Example 3 Double-sandwich ELISA detection of Aβ expression in HEK cells and 20E2 cells
[0045] HEK cells and 20E2 cells were cultured by conventional methods. Cloning the gene sequence of miR-7C (Let-7c), has-miR-99a (C99) into the p-super expression vector, constructing the expression plasmid of miR-7C, miR-99a, wherein has-miR-99a nucleoside The acid sequence is as shown in SEQ ID NO:2, specifically:
[0046] CCAUUGGCAUAAACCCGUAGAUCCGAUCUUGUGGUGAAGUGGACCGCACAAGCUCGCUUCUAUGGGUCUGUGUGUCAGUGUG. The plasmid was transfected into the cells according to the instructions of Lipofectamine plus, specifically: the constructed miR-7C expression plasmid and APP were co-transfected into HEK293 cells, the miR-7C and miR-99a expression plasmids were co-transfected into HEK293 cells, and miR-7C expression plasmids were co-transfected into HEK293 cells. The -7C expression plasmid was transfected into 20E2 cells alone. After the plasmid was transfected into the cells and cultured for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com