An Efficient Yeast Chromosome Fusion Method
A chromosome and yeast technology, applied in the field of efficient yeast chromosome fusion, can solve the problems of difficult DNA separation, limited application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0135] Example 1 , two chromosomal fusions
[0136] Using Saccharomyces cerevisiae BY4742 as the starting strain, ChrVI (270kbp) and ChrI (230kbp), ChrIX (440kbp) and ChrII (813kbp) were selected for fusion (Table 2).
[0137] Table 2. Two chromosome fusion information
[0138]
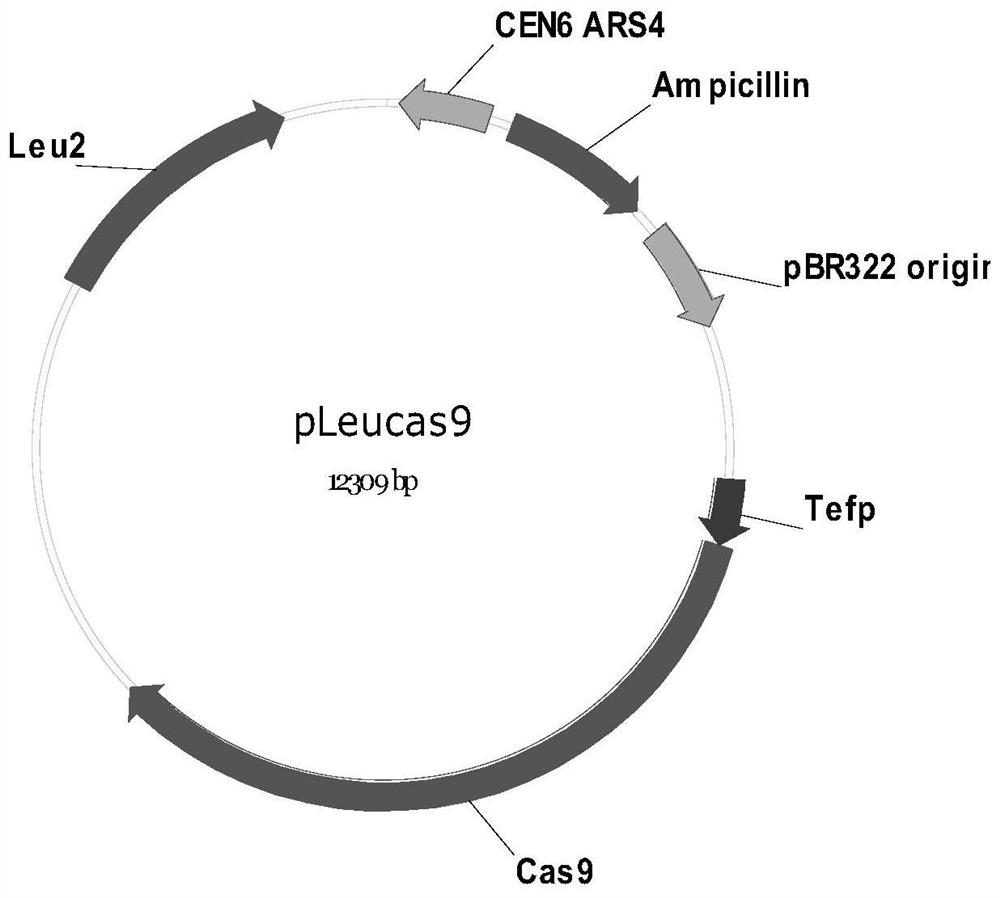
[0139] The Cas9 expression plasmid pleucas9 was transformed into BY4742 yeast cells to obtain strain BY4742 / pleucas9. Plasmid pleucas9 contains the promoter Tef1 to allow constitutive expression of Cas9 in yeast. The plasmid contains the yeast replication region (CEN6ARS4) and the selection marker (LEU2), which is single-copy replication in yeast; also contains the E. coli replication region (derived from pBR322) and the selection marker ampicillin (Ampicillin), which is high in E. coli Copy copy.
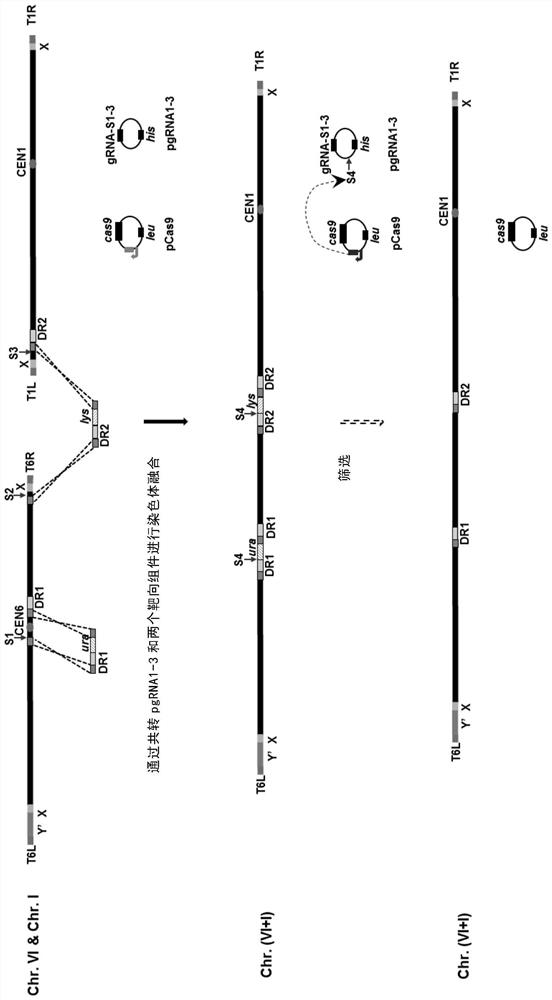
[0140] When ChrVI and ChrI are fused, the ChrVI centromere is knocked out, and 148410-148459bp, 148725-148774bp, 148775-149002bp on ChrVI are selected as the left homology arm, right homology arm, ...
Embodiment 2
[0144] Example 2 , three chromosome fusions
[0145] In this example, Saccharomyces cerevisiae BY4742 was used as the starting strain, and ChrVI (270kbp), ChrI (230kbp) and ChrII (813kbp) were selected for fusion, as shown in Table 3.
[0146] Table 3. Three chromosome fusion information
[0147]
[0148] ChrVI (270 kbp), ChrI (230 kbp) and ChrII (813 kbp) were fused to knock out the centromere of ChrVI and ChrII. Select 148410-148459bp, 148725-148774bp, 148775-149002bp on ChrVI as the left homology arm, right homology arm and forward repeat sequence of centromere knockout; 269212-269615bp as the left homology arm of the ChrVI-I fusion component . 2893-3294bp and 3295-3519bp on ChrI serve as the right homology arm and forward repeat of ChrVI-I fusion assembly; 202775-203182bp serve as the left homology arm of ChrI-II fusion assembly. 8680-9089bp and 9090-9359bp on ChrII serve as the right homology arm and forward repeat of the ChrI-II fusion component. Select 237974-2...
Embodiment 3
[0152] Example 3 , two sets of two chromosomal fusions
[0153] In this example, Saccharomyces cerevisiae BY4742 was used as the starting strain, and ChrVI (270kbp) and ChrI (230kbp), ChrIX (440kbp) and ChrII (813kbp) were simultaneously fused (Table 4).
[0154] Table 4. Two sets of chromosome fusion information
[0155]
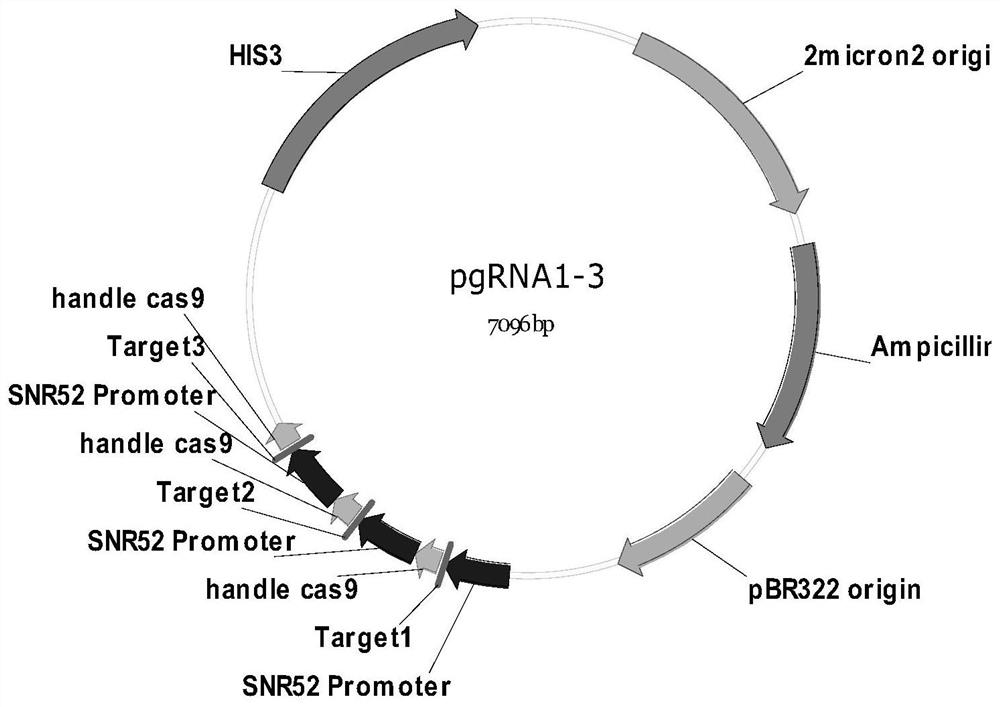
[0156] When ChrVI (270kbp) and ChrI (230kbp), ChrIX (440kbp) and ChrII (810kbp) are fused at the same time, pgRNA1-6, ChrVI centromere knockout module (URA3), ChrVI-I fusion module (URA3), ChrIX The centromeric knockout module (LYS2) and the ChrIX-II fusion module (LYS2) were transferred into BY4742 / pleucas9 using the protoplast fusion transformation method described in Example 1 (see Example 1 for the selection of homology arms).
[0157] Preliminary verification was carried out according to the colony PCR method described in Example 1 (see Example 1 for verification primers), two experiments were carried out, the number of fusion transformants was 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com