Fast detection method of cell line CRISPR/Cas9 gene knock-out
A technology for gene knockout and detection method, applied in the field of genetic engineering, can solve the problems of time-consuming and labor-intensive, expensive enzyme, narrow scope of application, etc., and achieve the effect of saving economy and time cost, high sensitivity and precision, and accurate positioning results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
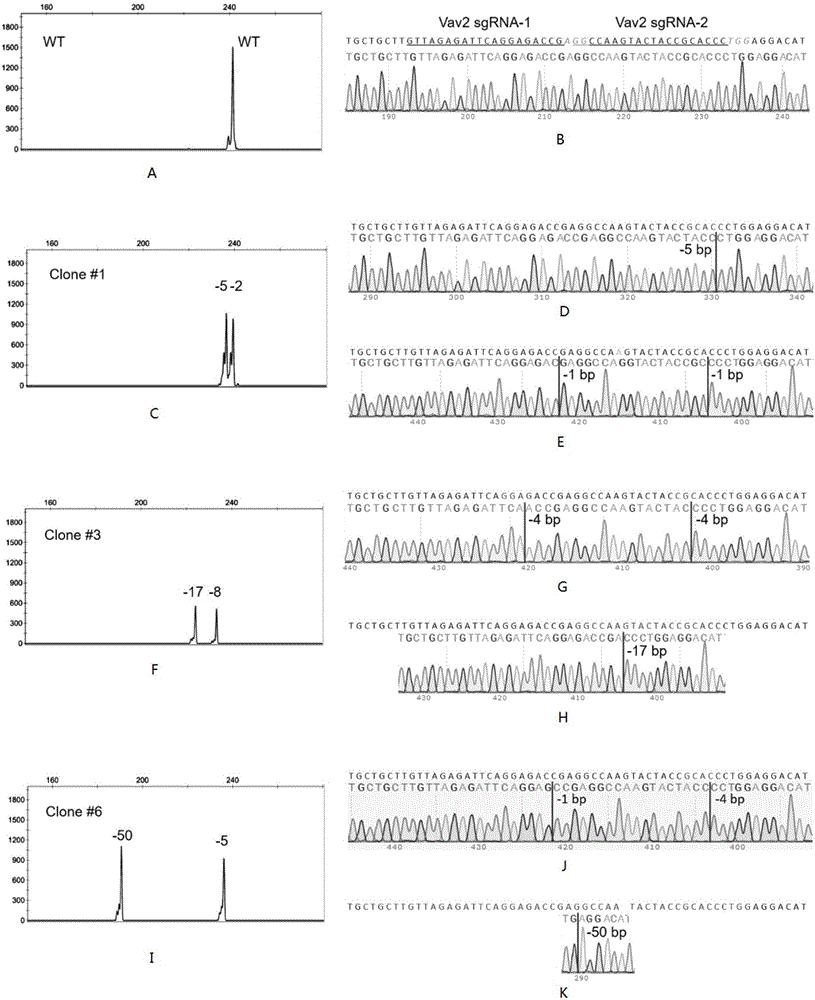
[0054] (1) Design a specific targeting site for the mouse Vav2 gene, construct a vector, transfect the cell line RAW264.7, and obtain GFP-positive single cells by flow cytometry: the mouse Vav2 gene ID is MGI: 102718 (MGI mouse gene database), the targeting site is 5'-GGCCAAGTACTACCGCACCCTGG-3' (SEQ ID NO.1) and 5'-GTTAGAGATTCAGGAGACCGAGG-3' (SEQ ID NO.2) in the Exon6 (ENSMUSE00001307648) segment, The vector backbone used pX458 (purchased from Addgene). After the vector was constructed, the cell line RAW264.7 was transfected with liposomes, and GFP-positive single cells were obtained by sorting with flow cytometry (BD fusion) after 40-80 hours. into a 96-well cell culture plate;
[0055] (2) Expand the culture of GFP-positive single cells, take part of the cells and directly lyse to obtain a DNA solution for subsequent PCR amplification: take a small amount of cells (do not need to quantify, about 100-1000), centrifugal force 350g, centrifuge Time 5min, discard the supernatan...
Embodiment 2
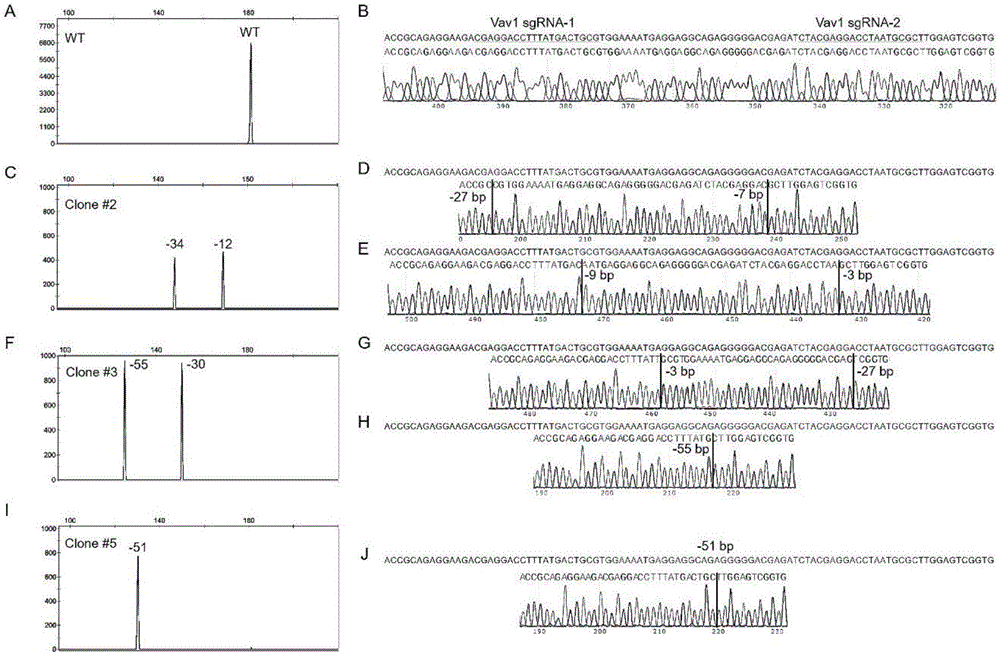
[0079] (1) Design a specific targeting site for the mouse Vav1 gene, construct a vector, transfect the cell line B16 and sort by flow cytometry, and finally obtain GFP-positive single cells: the mouse Vav1 gene ID is MGI: 98923 (MGI mouse gene database, Transcript ID: ENSMUST00000005889), the targeting site is (SEQ ID NO.20) in the segment of exon5 (exon ID: ENSMUSE00000138941): 5'-CTACGAGGACCTAATGCGCT TGG-3' and (SEQ ID NO. 21): 5'-CGAGGACCTTTTATGACTGCG TGG-3', the vector backbone uses pX458 (purchased from Addgene), after the vector is constructed, the cell line B16 is transfected by liposome, and after 40-72 hours, it is passed through the flow cytometer (BDfusion ) Sorting the GFP-positive single cells to a 96-well cell culture plate;
[0080](2) Expand the culture of GFP-positive single cells, take part of the cells and directly lyse to obtain a DNA solution for subsequent PCR amplification: take a small amount of cells (do not need to quantify, about 100-1000), centrifug...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com