Bacillus circulans chitoanase as well as preparation method and application thereof
A technology of Bacillus circulans and chitosanase, which is applied in the field of chitosanase, can solve the problems of large amount of enzyme used, increased production cost of chitosan oligosaccharide, and low ratio, and achieve huge application potential, improved efficiency, and high efficiency The effect of secretory expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Codon optimization and total gene synthesis of embodiment 1 chitosanase gene
[0033] Under the premise of not changing the amino acid sequence, the Bacillus circulans chitosanase (GH46 family) was artificially optimized using the preferred codons of Pichia pastoris (as shown in the sequence SEQ ID NO.1, GenBank accession number: BAA01474. 2) The coding gene, see SEQ ID NO.2 for the specific nucleotide sequence. The optimized nucleotide sequence and the original potential chitosanase coding gene sequence, as shown in the sequence SEQ ID NO.3, GenBank accession number: D10624.2 (569-1474), have the highest homology of 76%. The optimized gene sequence was entrusted to Sangong for full synthesis, and the synthesized gene sequence was named chitosanase gene bccsn.
Embodiment 2
[0034] The expression vector construction of embodiment 2 chitosanase gene bccsn
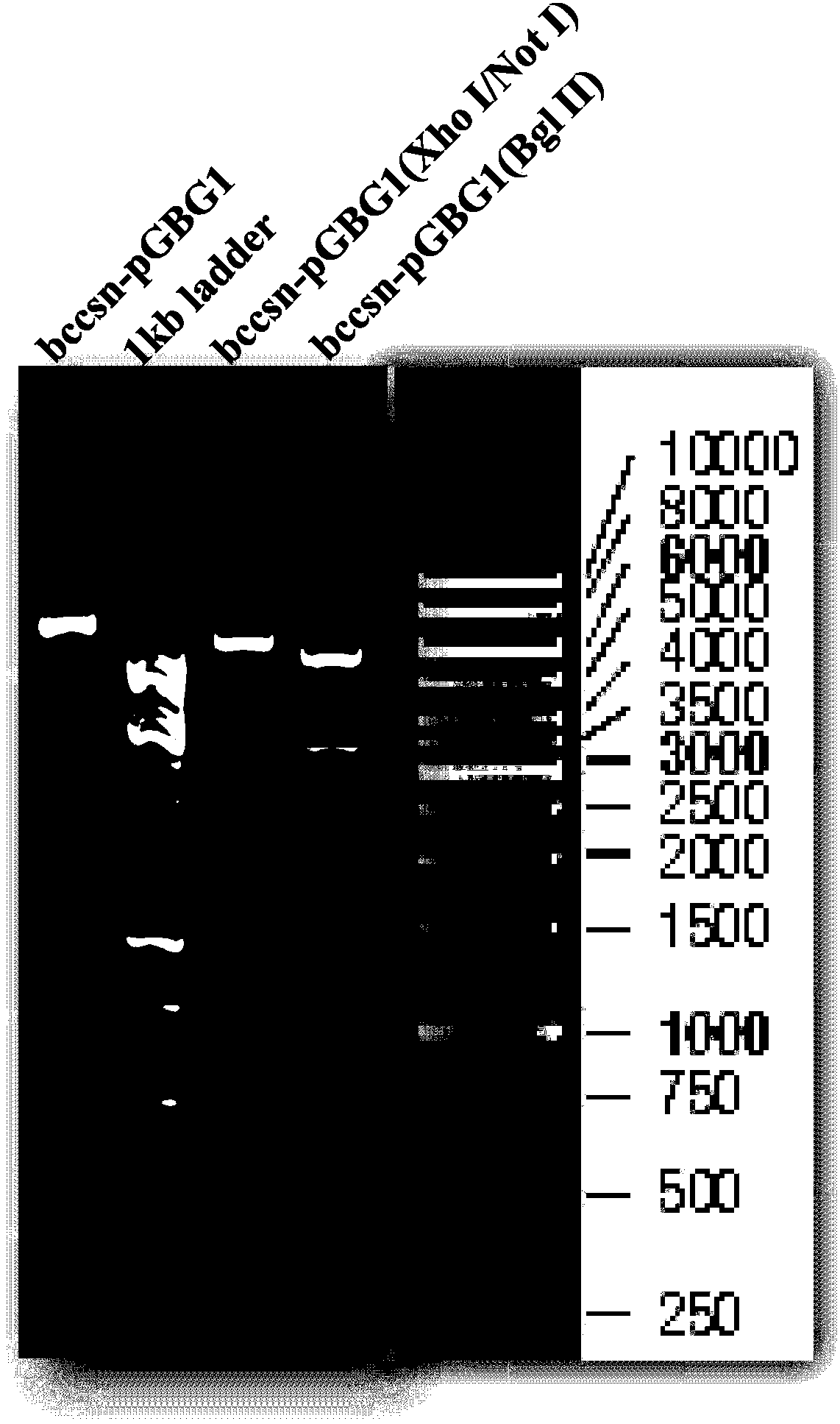
[0035] First, use restriction endonucleases Xho I and Not I to carry out double enzyme digestion on the cloning vector containing chitosanase gene bccsn to obtain the target gene fragment, and use the same endonuclease to carry out double enzyme digestion on the expression vector pGBG1, and recover large fragments. The two recovered products were connected to obtain a recombinant vector named bccsn-pGBG1. In order to confirm that the target chitosanase gene has been constructed into the vector, we respectively use Xho I / Not I and Bgl II to carry out double digestion and single digestion of the recombinant vector, and perform agarose gel electrophoresis on the product, the results are as follows figure 1 Shown: After double digestion, the target gene fragment appeared between 750bp and 1000bp, which was consistent with the 813bp fragment of bccsn; after Bgl II digestion, the expected two fragmen...
Embodiment 3
[0036] Example 3 The screening of chitosanase Pichia engineering bacteria and the preparation of chitosanase
[0037] After the obtained recombinant plasmid bccsn-pGBG1 was linearized by the restriction endonuclease BglII, gel electrophoresis was used to separate and excise the nucleotide fragment containing the target gene (such as figure 1 shown in the larger fragment), electroporation introduced into Pichia pastoris GS115, and the recombinant obtained by screening on the histidine auxotrophic MD plate was spread on the BMMY agar plate containing colloidal chitosan (0.5%) for cultivation , from which the monoclonal strain with the largest hydrolytic circle was screened out. A single colony of the screened monoclonal strain was inoculated in 200 mL of BMGY medium, cultured at 30°C and 250 rpm for 48 hours, the supernatant was discarded by centrifugation, and an equal amount of BMMY was added to induce expression. After 24 hours, methanol was added to a final concentration of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com