Method for extracting microbial metagenome DNA from intestinal tract content
A technology of metagenomic and content, applied in the field of extraction of microbial metagenomic DNA, can solve problems such as cracking and inability to obtain species abundance, and achieve the effects of reducing degradation, reducing species preference, and reducing pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
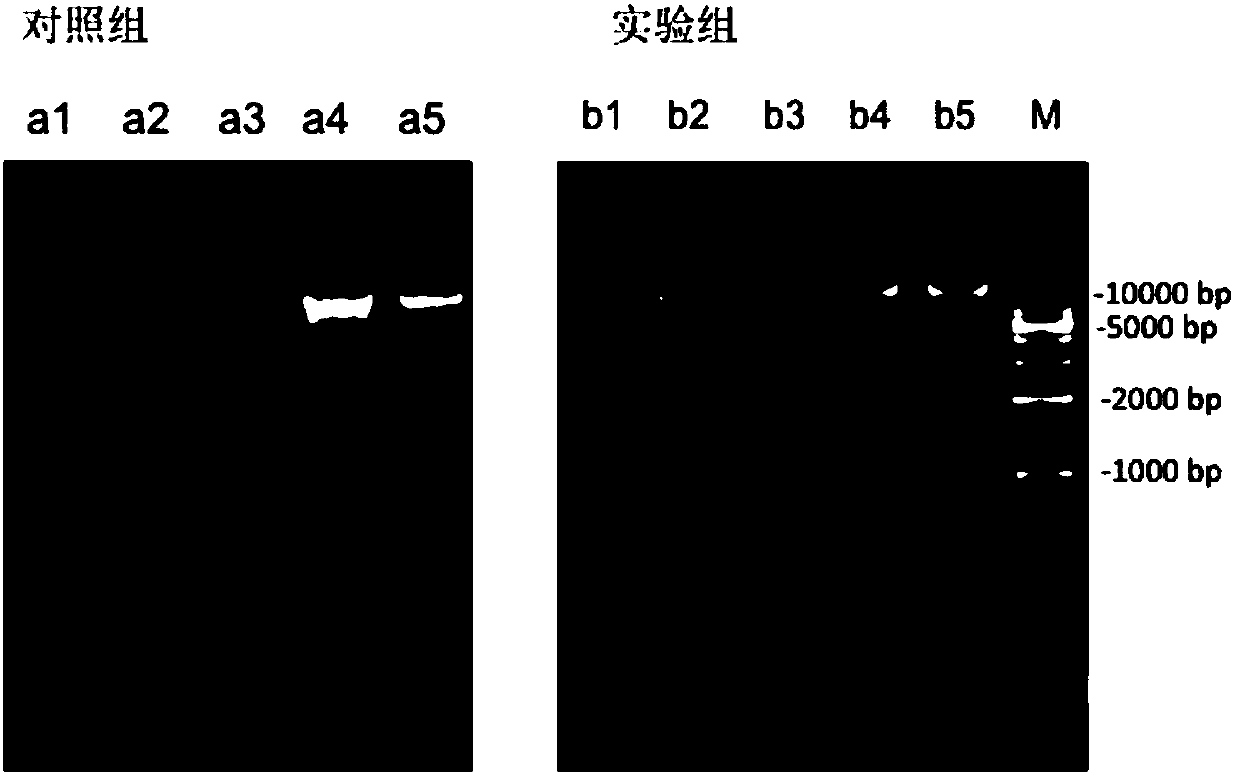
[0045] Experimental example 1. Extracting microbial metagenomic DNA from chicken intestinal contents
[0046] 1. Collection of intestinal contents of chicken intestine duodenum, jejunum, ileum, cecum, colorectal
[0047] Various fresh chicken intestinal contents were obtained by dissection, and 2 times volume of the chicken intestinal contents were added with 4°C pre-cooled normal saline containing 0.1% Tween80, and the samples were shaken and mixed thoroughly to obtain contents samples.
[0048] Take about 5-10g samples of duodenal contents, about 10-20g samples of jejunum contents, and about 10-20g samples of ileum contents into centrifuge tubes for subsequent steps; take about 0.5-1.0g samples of cecal contents, about 0.5 -1.0g sample of colorectal contents to centrifuge tube for subsequent steps.
[0049] 2. Extract microbial metagenomic DNA from chicken intestinal contents
[0050] 1. Differential centrifugation to wash and enrich intestinal microbial cells
[0051] 1) Centrifuge a...
Embodiment 2
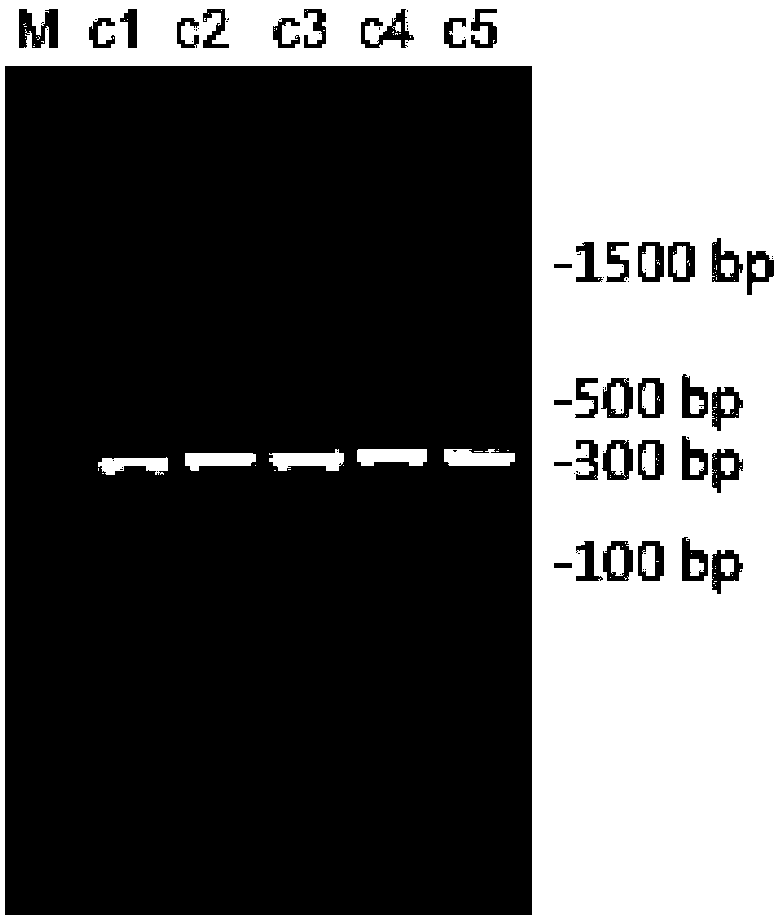
[0094] Example 2: Different lysis and fragmentation methods and analysis of microorganisms belonging to metagenomic DNA obtained
[0095] In this embodiment, a fresh chicken feces sample is used as a sample.
[0096] 1. Extract metagenomic DNA
[0097] 1. Differential centrifugation to wash and enrich microbial cells
[0098] Take 2 g of fresh chicken feces sample, wash and enrich the microbial cells according to the differential centrifugation in 1 of Experimental Example 12 to obtain microbial cells.
[0099] 2. The lysis of microbial cells
[0100] Divide the microbial cells obtained in the above 1 into 4 groups and lyse and break them according to the following 4 methods, specifically, they are divided into 4 2mL centrifuge tubes:
[0101] Method 1: Use 800uL cell lysate (QIAGEN company ASL buffer), lyse at 70℃ for 15min, centrifuge at 19000g for 5min, and collect the supernatant;
[0102] Method 2: Use 800uL cell lysate (QIAGEN ASL buffer), lyse at 95℃ for 15min, centrifuge at 19000g ...
experiment example 3
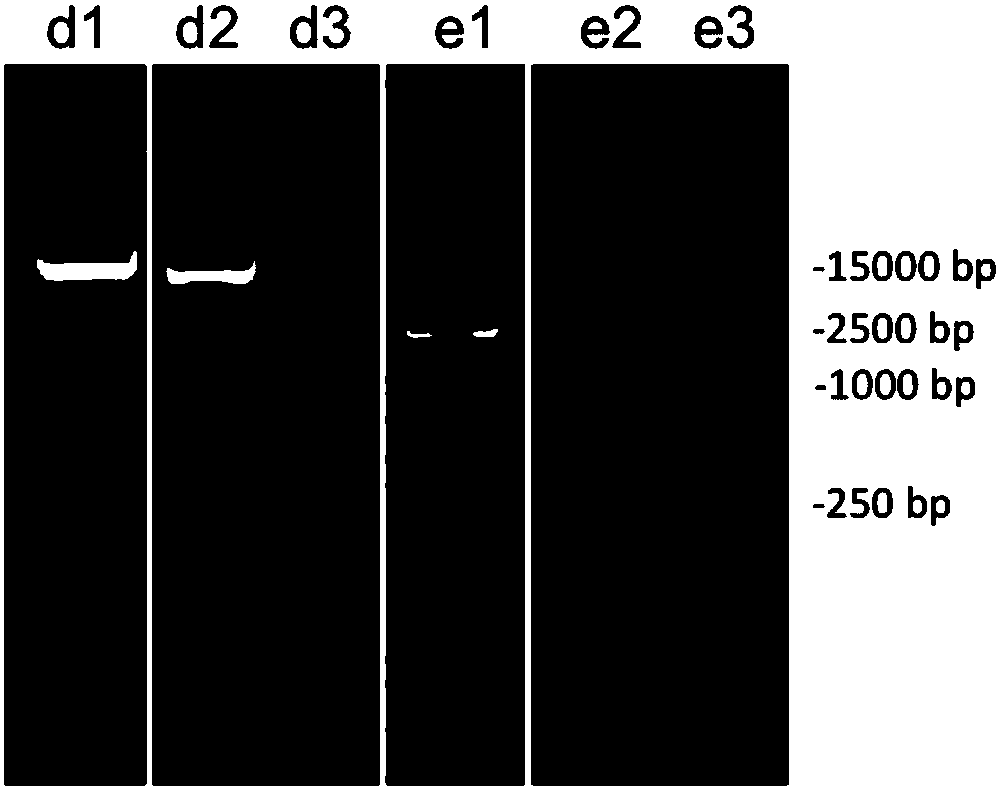
[0113] Experimental example 3: Exploring the importance of differential centrifugal washing steps
[0114] 1. Extract metagenomic DNA
[0115] 1. Collect the washing solution of the ileal contents that have not been washed by differential centrifugation
[0116] Take 0.5g sample of chicken intestinal ileum content, add 1mL of 4℃ pre-cooled normal saline (containing 0.1% Tween80), shake well to mix the sample, centrifuge at 4℃ and 19000g for 15min at high speed, sediment the chyme and Microbial cells are collected from the washing liquid of the ileal contents.
[0117] 2. The metagenomic DNA sample from the cecal contents obtained in Experimental Example 1 was diluted to a concentration of 50ng / uL to become the DNA sample d1.
[0118] The DNA molecular weight standard (Tiangen Biochemical Technology Co., Ltd., D15000) with a concentration of 50ng / uL is the DNA sample e1. Take 5uL DNA samples d1 and e1, mix them with 5uL physiological saline (containing 0.1% Tween80), and react at room ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com